Explore the hospital effect on thrombolysis use, using SHAP values and SHAP main effects from a single XGBoost model#
Research question: What’s the difference between the SHAP main effect (and SHAP value) for patients that attend a hospital, with the SHAP main effect (and SHAP value) for patients that do not attend a hospital
Combining the results from the previous two notebooks: 03b and 03c
Plain English summary#
When fitting a machine learning model to data to make a prediction, it is now possible, with the use of the SHAP library, to allocate contributions of the prediction onto the feature values. This means that we can now turn these black box methods into transparent models and describe what the model used to obtain it’s prediction.
SHAP values are calculated for each feature of each instance for a fitted model. In addition there is the SHAP base value which is the same value for all of the instances. The base value represents the models best guess for any instance without any extra knowledge about the instance (this can also be thought of as the “expected value”). It is possible to obtain the models prediction of an instance by taking the sum of the SHAP base value and each of the SHAP values for the features. This allows the prediction from a model to be transparant, and we can rank the features by their importance in determining the prediction for each instance.
The SHAP values for each feature are comprised of the feature’s main effect (what is due to the feature value, the standalone effect) and all of the pairwise interaction effects with each of the other features (a value per feature pairings).
In two other detailed notebooks (see GitHub repo for these notebooks: https://github.com/samuel-book/samuel_shap_paper_1), we looked at SHAP values (03b) and SHAP main effect (3c) for the one-hot encoded hospital features. We looked at the values for patients that attend the hospital, and for those that do not.
Here we load the SHAP values (03a) and SHAP interaction values (03c) for the single XGBoost model fitted to all the data (no test set), and analyse the SHAP values and SHAP main effect values for each of the one-hot encoded hospital features.
SHAP values are in the same units as the model output (for XGBoost these are in log odds).
Model and data#
Using the XGBoost model trained on all of the data (no test set used) from notebook 03a_xgb_combined_shap_key_features.ipynb. The 10 features in the model are:
Arrival-to-scan time: Time from arrival at hospital to scan (mins)
Infarction: Stroke type (1 = infarction, 0 = haemorrhage)
Stroke severity: Stroke severity (NIHSS) on arrival
Precise onset time: Onset time type (1 = precise, 0 = best estimate)
Prior disability level: Disability level (modified Rankin Scale) before stroke
Use of AF anticoagulents: Use of atrial fibrillation anticoagulant (0 = No, 1 = Yes)
Onset-to-arrival time: Time from onset of stroke to arrival at hospital (mins)
Onset during sleep: Did stroke occur in sleep?
Age: Age (as middle of 5 year age bands)
Stroke team: Represented as one-hot encoded features
And one target feature:
Thrombolysis: Did the patient recieve thrombolysis (0 = No, 1 = Yes)
Aims#
Using XGBoost model fitted on all the data (no test set) using 10 features
Using SHAP values and SHAP interaction values previously calculated
Understand the relationship between SHAP values and SHAP main effect values for the hospital one-hot encoded features
Observations#
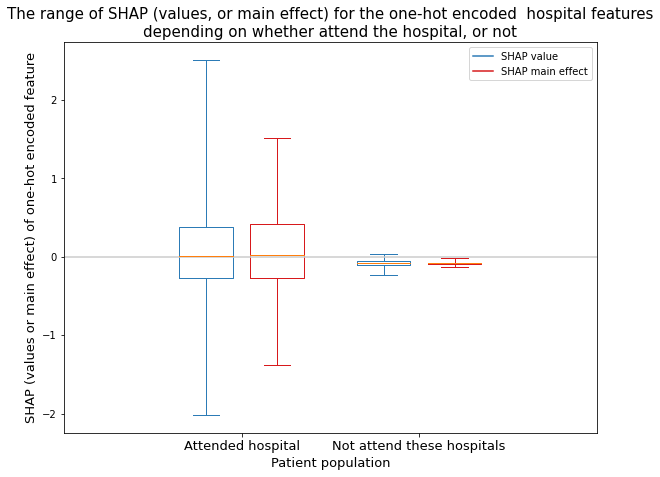
Both of the SHAP values and SHAP main effect values for the one-hot encoded hospital features are very dependent on whether the instance attended the hospital or not
For the instances that attend a hospital, the SHAP main effect values are a narrower range than the equivalent SHAP values (due to the SHAP values being made up of the main effect and the pair-wise feature interactions).
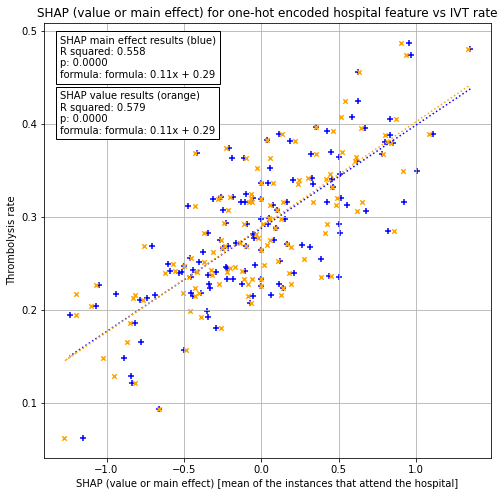
56% of the variability in hospital thrombolysis rate can be explained by the SHAP main effect value for the one-hot encoded hospital feature (the mean of those instances that attend the hospital).
58% of the variability in hospital thrombolysis rate can be explained by the SHAP main effect value for the one-hot encoded hospital feature (the mean of those instances that attend the hospital).
Import libraries#
# Turn warnings off to keep notebook tidy
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
# Import machine learning methods
from xgboost import XGBClassifier
# Import shap for shapley values
import shap
from scipy import stats
import os
import pickle
import math # to .floor and .ceil
# So can take deep copy
import copy
from os.path import exists
import json
Set filenames#
# Set up strings (describing the model) to use in filenames
number_of_features_to_use = 10
model_text = f'xgb_{number_of_features_to_use}_features'
notebook = '03d'
Create output folders if needed#
path = './saved_models'
if not os.path.exists(path):
os.makedirs(path)
path = './output'
if not os.path.exists(path):
os.makedirs(path)
path = './predictions'
if not os.path.exists(path):
os.makedirs(path)
Read in JSON file#
Contains a dictionary for plain English feature names for the 10 features selected in the model. Use these as the column titles in the DataFrame.
with open("./output/01_feature_name_dict.json") as json_file:
dict_feature_name = json.load(json_file)
Load data#
Data has previously been split into 5 stratified k-fold splits.
The model used in this notebook is fitted using all of the data (rather than train/test splits used to assess accuracy). We will join up all of the test data (by definition, each instance exists only once across all of the 5 test sets)
data_loc = '../data/kfold_5fold/'
# Initialise empty list
test_data_kfold = []
# Read in the names of the selected features for the model
key_features = pd.read_csv('./output/01_feature_selection.csv')
key_features = list(key_features['feature'])[:number_of_features_to_use]
# And add the target feature name: S2Thrombolysis
key_features.append('S2Thrombolysis')
# For each k-fold split
for i in range(5):
# Read in test set, restrict to chosen features, rename titles, & store
test = pd.read_csv(data_loc + 'test_{0}.csv'.format(i))
test = test[key_features]
test.rename(columns=dict_feature_name, inplace=True)
test_data_kfold.append(test)
# Join all the test sets. Set "ignore_index = True" to reset the index in the
# new dataframe (to run from 0 to n-1), otherwise get duplicate index values
data = pd.concat(test_data_kfold, ignore_index=True)
Get list of feature names#
feature_names = list(test_data_kfold[0])
Edit data#
Divide into X (features) and y (labels)#
We will separate out our features (the data we use to make a prediction) from our label (what we are trying to predict).
By convention our features are called X (usually upper case to denote multiple features), and the label (thrombolysis or not) y.
X = data.drop('Thrombolysis', axis=1)
y = data['Thrombolysis']
Average thromboylsis (this is the expected outcome of each patient, without knowing anything about the patient)
print (f'Average treatment: {round(y.mean(),2)}')
Average treatment: 0.3
One-hot encode hospital feature#
# Keep copy of original, with 'Stroke team' not one-hot encoded
X_combined = X.copy(deep=True)
# One-hot encode 'Stroke team'
X_hosp = pd.get_dummies(X['Stroke team'], prefix = 'team')
X = pd.concat([X, X_hosp], axis=1)
X.drop('Stroke team', axis=1, inplace=True)
XGBoost model#
An XGBoost model was trained on the full dataset (rather than train/test splits used to assess accuracy) in notebook 03a. These models are saved f’./saved_models/03a_{model_text}.p’.
Get SHAP values#
SHAP values give the contribution that each feature has on the models prediction, per instance. A SHAP value is returned for each feature, for each instance.
‘Raw’ SHAP values from XGBoost model are log odds ratios.
In notebook 03a we used the TreeExplainer from the shap library (https://shap.readthedocs.io/en/latest/index.html) to calculate the SHAP values. This is a fast and exact method to estimate SHAP values for tree models and ensembles of tree.
Load SHAP values from pickle (calculated in notebook 03a).
filename = f'./output/03a_{model_text}_shap_values_extended.p'
# Load SHAP values
with open(filename, 'rb') as filehandler:
shap_values_extended = pickle.load(filehandler)
The explainer returns the base value which is the same value for all instances [shap_value.base_values], the shap values per feature [shap_value.values]. It also returns the feature dataset values [shap_values.data]. You can (sometimes!) access the feature names from the explainer [explainer.data_feature_names].
Let’s take a look at the data held for the first instance:
.values has the SHAP value for each of the four features.
.base_values has the best guess value without knowing anything about the instance.
.data has each of the feature values
shap_values_extended[0]
.values =
array([ 7.56502628e-01, 4.88319576e-01, 1.10273695e+00, 4.40223962e-01,
4.98975307e-01, 2.02812225e-01, -2.62601525e-01, 3.69181409e-02,
2.30234995e-01, 3.20923195e-04, -6.03703642e-03, -7.17267860e-04,
0.00000000e+00, -4.91688668e-04, 1.20360847e-03, 1.77788886e-03,
-4.34198463e-03, -2.76021136e-04, -2.79896380e-03, 3.52182891e-03,
-2.73969141e-04, 8.53505917e-03, -5.28220041e-03, -8.25227005e-04,
6.20208494e-03, 6.92215608e-03, -6.32244349e-03, -3.35367222e-04,
7.81939551e-03, -4.71850217e-06, -4.25534381e-05, 6.48253039e-03,
8.43156071e-04, -6.28353562e-04, -1.25156669e-02, -7.92063680e-03,
-1.99409085e-03, -5.05548809e-03, -3.90118686e-03, 1.30317558e-03,
0.00000000e+00, -6.48246554e-04, 1.19629130e-03, 8.26304778e-04,
1.28053436e-02, 2.55714403e-03, -3.20375757e-03, 4.23251512e-03,
-7.19791185e-03, 4.02670400e-03, 3.75146419e-03, 8.31848301e-04,
3.45067866e-03, 3.92199913e-03, 1.05317042e-04, 9.53648426e-03,
2.34490633e-03, -1.05699897e-03, -7.75758363e-03, 1.09220366e-03,
0.00000000e+00, 5.15310653e-03, -1.28013985e-02, 7.28079583e-03,
1.98326469e-03, 2.40865033e-04, 6.70310017e-03, 1.18395500e-03,
8.82393331e-04, 2.83133984e-03, 2.06021918e-03, -8.22589453e-03,
5.11038816e-03, -3.25066457e-03, -1.15480982e-02, 9.36000666e-04,
2.03671609e-03, -2.02708528e-03, -5.31264208e-03, 2.42300611e-03,
3.16989725e-04, 3.67143471e-03, -5.07418578e-03, -2.68517504e-03,
3.30522249e-04, -6.63852552e-03, -1.63316587e-03, 1.75383547e-03,
-4.12231544e-03, 1.20234396e-03, -6.25999365e-03, 0.00000000e+00,
-1.00428164e-02, -1.76329317e-03, 3.12283775e-03, 4.69124934e-04,
-1.22163491e-03, 2.30912305e-03, 9.43152234e-04, 1.22348359e-03,
5.29656609e-05, -9.66969132e-03, 3.44762520e-06, -5.46710449e-04,
4.26546484e-03, 4.65912558e-03, 1.30611981e-04, -2.89813057e-03,
-2.98934802e-03, -1.47398096e-03, -2.31104009e-02, -3.47609469e-03,
-5.73671749e-03, 3.97895870e-04, 9.02379164e-04, -5.86819311e-04,
-1.79305486e-03, -5.28960605e-04, -2.10736915e-02, 3.50499223e-03,
-2.04754542e-04, -2.86527374e-03, 1.52953295e-03, -3.72403156e-06,
1.42271689e-03, -2.97368824e-04, 5.06201060e-04, 5.77868486e-04,
-1.44459037e-02, 3.30785802e-03, -2.37809308e-03, -7.35214865e-03,
3.36531224e-03, 1.49519066e-03, -2.46208580e-03, 7.20066251e-04,
6.83251710e-04, -1.92034326e-03, 2.39002449e-03, -8.96935933e-04,
4.85493708e-03], dtype=float32)
.base_values =
-1.1629876
.data =
array([ 17. , 1. , 14. , 1. , 0. , 0. , 186. , 0. , 47.5,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 1. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. ])
There is one of these for each instance.
shap_values_extended.shape
(88792, 141)
Format the SHAP values data#
Features are in the same order in shap_values as they are in the original dataset. Use this fact to extract the SHAP values for the one-hot encoded hospital features.
Create a dataframe containing the SHAP values: an instance per row, and a one-hot encoded hospital feature per column.
# Get list of hospital one hot encoded column titles
hospital_names_ohe = X.filter(regex='^team',axis=1).columns
n_hospitals = len(hospital_names_ohe)
# Get list of hospital names without the prefix "team_"
hospital_names = [h[5:] for h in hospital_names_ohe]
# Create list of column index for these hospital column titles
hospital_columns_index = [X.columns.get_loc(col) for col in hospital_names_ohe]
# Use this index list to access the hosptial shap values (as array)
hosp_shap_values = shap_values_extended.values[:,hospital_columns_index]
# Put in dataframe with hospital as column title
df_hosp_shap_values = pd.DataFrame(hosp_shap_values, columns = hospital_names)
Add these columns:
Stroke team instance attended
contribution from all the hospital features
contribution from attending the hospital
contribution from not attending the rest
# Include Stroke team that each instance attended
df_hosp_shap_values["Stroke team"] = X_combined["Stroke team"].values
# Store the sum of the SHAP values (for all of the hospital features)
df_hosp_shap_values["all_stroke_teams"] = df_hosp_shap_values.sum(axis=1)
# Initialise list for 1) SHAP value for attended hospital, &
# 2) SHAP value for the sum of the rest of the hospitals
shap_values_attended_hospital = []
shap_values_not_attend_these_hospitals = []
# For each patient
for index, row in df_hosp_shap_values.iterrows():
# Get stroke team attended
stroke_team = row["Stroke team"]
# Get SHAP value for the stroke team attended
shap_values_attended_hospital.append(row[stroke_team])
# Calculate sum of SHAP values for the stroke teams not attend
sum_rest = row["all_stroke_teams"] - row[stroke_team]
shap_values_not_attend_these_hospitals.append(sum_rest)
# Store two new columns in dataframe
df_hosp_shap_values["attended_stroke_team"] = shap_values_attended_hospital
df_hosp_shap_values["not_attended_stroke_teams"] = (
shap_values_not_attend_these_hospitals)
# View preview
df_hosp_shap_values.head()
| AGNOF1041H | AKCGO9726K | AOBTM3098N | APXEE8191H | ATDID5461S | BBXPQ0212O | BICAW1125K | BQZGT7491V | BXXZS5063A | CNBGF2713O | ... | YEXCH8391J | YPKYH1768F | YQMZV4284N | ZBVSO0975W | ZHCLE1578P | ZRRCV7012C | Stroke team | all_stroke_teams | attended_stroke_team | not_attended_stroke_teams | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.000321 | -0.006037 | -0.000717 | 0.0 | -0.000492 | 0.001204 | 0.001778 | -0.004342 | -0.000276 | -0.002799 | ... | 0.000720 | 0.000683 | -0.001920 | 0.002390 | -0.000897 | 0.004855 | TXHRP7672C | -0.084398 | -0.023110 | -0.061287 |
| 1 | 0.001207 | -0.002507 | 0.000081 | 0.0 | -0.001356 | 0.001547 | 0.003449 | -0.006348 | -0.000127 | -0.002576 | ... | 0.000148 | 0.000683 | -0.001825 | 0.000454 | -0.001445 | 0.003970 | SQGXB9559U | -0.760522 | -0.671515 | -0.089007 |
| 2 | 0.001492 | -0.015246 | 0.002635 | 0.0 | -0.000272 | 0.001399 | 0.002737 | -0.007848 | -0.000605 | -0.002614 | ... | 0.000135 | 0.000262 | -0.000928 | 0.001229 | -0.001506 | 0.003792 | LFPMM4706C | -1.204099 | -1.084834 | -0.119265 |
| 3 | 0.001255 | 0.002451 | 0.002644 | 0.0 | -0.001359 | 0.000008 | 0.003055 | -0.003141 | -0.000131 | -0.002352 | ... | 0.000705 | 0.000683 | -0.003016 | 0.003507 | -0.000939 | 0.002133 | MHMYL4920B | 0.684426 | 0.737010 | -0.052583 |
| 4 | 0.006129 | -0.004731 | 0.002644 | 0.0 | -0.001359 | 0.000302 | 0.002677 | -0.004384 | 0.000272 | -0.002281 | ... | 0.000892 | 0.000010 | -0.001976 | 0.001467 | -0.000799 | 0.002244 | EQZZZ5658G | -0.088429 | 0.028813 | -0.117242 |
5 rows × 136 columns
Get SHAP interaction values#
A SHAP interaction value is returned for each pair of features (including with itself, which is known as the main effect), for each instance. The SHAP value for a feature is the sum of it’s pair-wise feature interactions.
We used the TreeExplainer (from the shap library: https://shap.readthedocs.io/en/latest/index.html) to calculate the SHAP interaction values (notebook 03c).
‘Raw’ SHAP interaction values from XGBoost model are log odds ratios.
Load from pickle (from notebook 03c).
%%time
filename = f'./output/03c_{model_text}_shap_interactions.p'
# Load SHAP interaction
with open(filename, 'rb') as filehandler:
shap_interactions = pickle.load(filehandler)
CPU times: user 38.5 ms, sys: 3 s, total: 3.04 s
Wall time: 4.14 s
SHAP interaction values have a matrix of values (per pair of features) per instance.
In this case, each of the 88792 instances has a 139x139 matrix of SHAP interaction values (with the SHAP main effect on the diagonal positions).
shap_interactions.shape
(88792, 141, 141)
Show SHAP interation matrix (with main effect on the diagonal positions) for the first instance. Notice how the SHAP interation for pairs of features are symmetrical across the diagonal.
shap_interactions[0]
array([[ 6.76276624e-01, 4.08733338e-02, 1.05852485e-02, ...,
1.75262336e-04, 1.55542511e-05, -3.98763688e-04],
[ 4.08733785e-02, 3.38386327e-01, 5.30318618e-02, ...,
0.00000000e+00, 0.00000000e+00, 6.54254109e-07],
[ 1.05856061e-02, 5.30319214e-02, 8.62581849e-01, ...,
1.82669377e-04, 3.62747931e-04, 8.75864644e-05],
...,
[ 1.75237656e-04, 0.00000000e+00, 1.82688236e-04, ...,
3.14869266e-03, 0.00000000e+00, 3.90200876e-06],
[ 1.55568123e-05, 0.00000000e+00, 3.62753868e-04, ...,
0.00000000e+00, -1.80786219e-03, 0.00000000e+00],
[-3.98725271e-04, 6.55651093e-07, 8.76188278e-05, ...,
3.90200876e-06, 0.00000000e+00, 3.13467532e-03]], dtype=float32)
Format the SHAP interaction data#
Features are in the same order in shap_interaction as they are in the original dataset. Use this fact to extract the SHAP main effect values for the one-hot encoded hospital features.
Create a dataframe containing the SHAP values: an instance per row, and a one-hot encoded hospital feature per column.
hosp_shap_main_effects = []
# Use index list to access the hosptial shap values (as array) in the loop below
for i in range(shap_interactions.shape[0]):
# Get the main effect value for each of the features
main_effects = np.diagonal(shap_interactions[i])
hosp_shap_main_effects.append(main_effects[hospital_columns_index])
# Put in dataframe with hospital as column title
df_hosp_shap_main_effects = pd.DataFrame(hosp_shap_main_effects,
columns = hospital_names)
Add these columns:
Stroke team instance attended
contribution from all the hospital features
contribution from attending the hospital
contribution from not attending the rest
# Include Stroke team that each instance attended
df_hosp_shap_main_effects["Stroke team"] = X_combined["Stroke team"].values
# Store the sum of the SHAP values (for all of the hospital features)
df_hosp_shap_main_effects["all_stroke_teams"] = (
df_hosp_shap_main_effects.sum(axis=1))
# Initialise lists for
# 1) SHAP value for attended hospital
# 2) SHAP value for the sum of the rest of the hospitals
shap_me_attended_hospital = []
shap_me_not_attend_these_hospitals = []
# For each patient
for index, row in df_hosp_shap_main_effects.iterrows():
# Get stroke team attended
stroke_team = row["Stroke team"]
# Get SHAP value for the stroke team attended
shap_me_attended_hospital.append(row[stroke_team])
# Calculate sum of SHAP values for the stroke teams not attend
sum_rest = row["all_stroke_teams"] - row[stroke_team]
shap_me_not_attend_these_hospitals.append(sum_rest)
# Store two new columns in dataframe
df_hosp_shap_main_effects["attended_stroke_team"] = shap_me_attended_hospital
df_hosp_shap_main_effects["not_attended_stroke_teams"] = (
shap_me_not_attend_these_hospitals)
# View preview
df_hosp_shap_main_effects.head()
| AGNOF1041H | AKCGO9726K | AOBTM3098N | APXEE8191H | ATDID5461S | BBXPQ0212O | BICAW1125K | BQZGT7491V | BXXZS5063A | CNBGF2713O | ... | YEXCH8391J | YPKYH1768F | YQMZV4284N | ZBVSO0975W | ZHCLE1578P | ZRRCV7012C | Stroke team | all_stroke_teams | attended_stroke_team | not_attended_stroke_teams | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.001171 | -0.007868 | 0.002866 | 0.0 | -0.000882 | 0.001910 | 0.002818 | -0.004191 | -0.000530 | -0.002555 | ... | 0.000343 | 0.000520 | -0.001912 | 0.003149 | -0.001808 | 0.003135 | TXHRP7672C | -0.280340 | -0.187852 | -0.092488 |
| 1 | 0.002057 | -0.011158 | 0.002381 | 0.0 | -0.000742 | 0.001681 | 0.002762 | -0.003978 | -0.000381 | -0.002564 | ... | 0.000422 | 0.000520 | -0.001846 | 0.002632 | -0.001676 | 0.004031 | SQGXB9559U | -0.668289 | -0.580359 | -0.087931 |
| 2 | 0.002480 | -0.009465 | 0.002425 | 0.0 | -0.001077 | 0.001824 | 0.002767 | -0.003166 | -0.000859 | -0.002645 | ... | 0.000321 | 0.000522 | -0.001428 | 0.003050 | -0.001737 | 0.004724 | LFPMM4706C | -1.278658 | -1.192582 | -0.086076 |
| 3 | 0.002105 | -0.009234 | 0.002305 | 0.0 | -0.000745 | 0.002322 | 0.002844 | -0.003517 | -0.000385 | -0.002717 | ... | 0.000328 | 0.000520 | -0.001788 | 0.002736 | -0.001850 | 0.003941 | MHMYL4920B | 0.910882 | 0.977001 | -0.066120 |
| 4 | 0.000452 | -0.008371 | 0.002305 | 0.0 | -0.000745 | 0.002055 | 0.002782 | -0.003513 | -0.000565 | -0.002647 | ... | 0.000326 | 0.000524 | -0.001853 | 0.002636 | -0.001710 | 0.004052 | EQZZZ5658G | -0.004142 | 0.070702 | -0.074844 |
5 rows × 136 columns
Create boxplot (range of SHAP values and SHAP main effect across 132 hospitals)#
Analyse the range of SHAP values and SHAP main effect values for the one-hot encoded hospital features. Show as two populations: the attended hospital, the sum of the hospitals not attended
To create a grouped boxplot, used code from https://stackoverflow.com/questions/16592222/matplotlib-group-boxplots
To define HEX colour codes use http://colorbrewer2.org/
def set_box_color(bp, color):
"""
bp [boxplot object]
color [Hex Color Codes]
"""
plt.setp(bp['boxes'], color=color)
plt.setp(bp['whiskers'], color=color)
plt.setp(bp['caps'], color=color)
plt.setp(bp['means'], color=color)
return()
fig = plt.figure(figsize=(8,6))
ax = fig.add_subplot(1,1,1)
ticks = ["Attended hospital", "Not attend these hospitals"]
# SHAP value for plot
# A list of two lists, the values in each list create a box on the plot
plot_data_sv = [shap_values_attended_hospital,
shap_values_not_attend_these_hospitals]
bp_sv = plt.boxplot(plot_data_sv,
positions=np.array(range(len(plot_data_sv)))*2.0-0.4,
sym='', whis=99999, widths=0.6)
set_box_color(bp_sv, '#2C7BB6')
# SHAP main effect for plot
# A list of two lists, the values in each list create a box on the plot
plot_data_me = [shap_me_attended_hospital, shap_me_not_attend_these_hospitals]
bp_me = plt.boxplot(plot_data_me,
positions=np.array(range(len(plot_data_me)))*2.0+0.4,
sym='', whis=99999, widths=0.6)
set_box_color(bp_me, '#D7191C')
# draw temporary red and blue lines and use them to create a legend
plt.plot([], c='#2C7BB6', label='SHAP value')
plt.plot([], c='#D7191C', label='SHAP main effect')
plt.legend()
# X axis
plt.xticks(range(0, len(ticks) * 2, 2), ticks, size=13)
plt.xlim(-2, len(ticks)*2)
plt.tight_layout()
# Add line at Shap = 0
ax.plot([plt.xlim()[0], plt.xlim()[1]], [0,0], c='0.8')
title = ("The range of SHAP (values, or main effect) for the one-hot encoded "
" hospital features\ndepending on whether attend the hospital, or not")
plt.title(title, size=15)
plt.ylabel('SHAP (values or main effect) of one-hot encoded feature', size=13)
plt.xlabel('Patient population', size=13)
plt.savefig(f'./output/{notebook}_{model_text}'
f'_hosp_shap_value_and_main_effect_attend_vs_notattend_boxplot.jpg',
dpi=300, bbox_inches='tight', pad_inches=0.2)
plt.show()
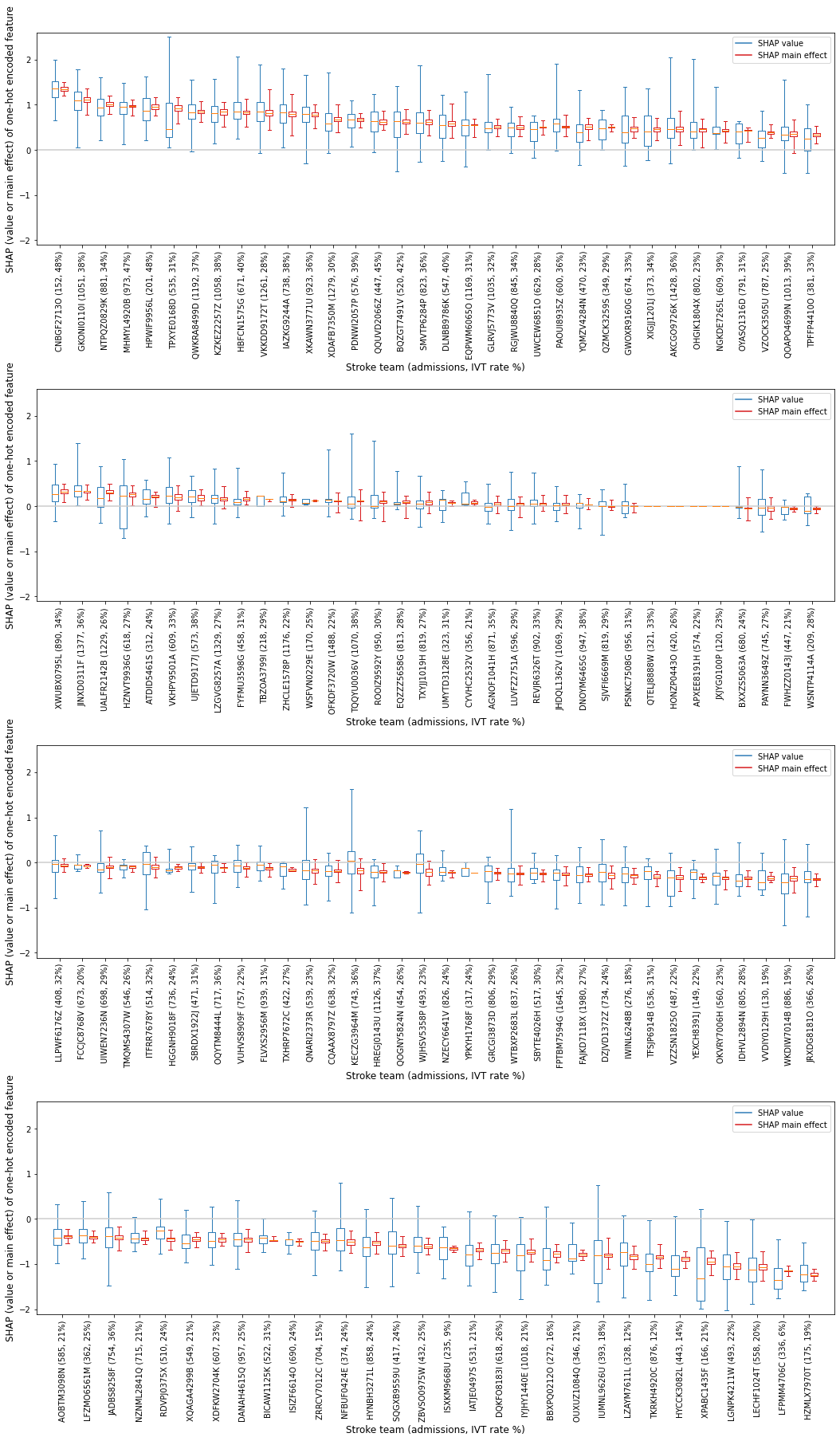
Create boxplot (range of SHAP values and SHAP main effect within each 132 hospitals)#
Create a boxplot to show the range of SHAP values and SHAP main effect values for each individual one-hot encoded hospital feature.
Show the SHAP value as two populations: 1) the group of instances that attend the hospital [black], and 2) the group of instances that do not attend the hosptial [orange].
Order the hospitals in descending order of mean SHAP value for the hospital the instance attended (so those that more often contribute to a yes-thrombolysis decision, through to those that most often contribute to a no-thrombolysis decision).
Firstly, to order the hospitals, create a dataframe containing the mean SHAP main effect, and mean SHAP values for each hosptial (for those instances that attended the hospital)
def shap_descriptive_stats(hospital_names, df_hosp_shap, prefix_text):
"""
For a set of columns (those contained in "hospital_names"), take only the
values which represented a hospital attended (using a mask from column
"stroke team". Return the descripive values of the IQR for the attended
hospital values (in a dataframe with hospital as row and each IQR as a
column.
hospital_names [list]: The list of unique hospital names
df_hosp_shap [DataFrame]: Contains a value per hospital (column) per
patient (row)
prefix_text [string]: Define what values are in the dataframe
return [DataFrame]: A hospital per row and each IQR as a column
"""
# Create list of values (one per hospital) for those instances that attend
# the hospital. Store the IQR for all the patients that attend the
# hospital
attend_stroketeam_min = []
attend_stroketeam_q1 = []
attend_stroketeam_mean = []
attend_stroketeam_q3 = []
attend_stroketeam_max = []
for h in hospital_names:
mask = df_hosp_shap['Stroke team'] == h
data_stroke_team = df_hosp_shap[h][mask]
q1, q3 = np.percentile(data_stroke_team, [25,75])
attend_stroketeam_min.append(data_stroke_team.min())
attend_stroketeam_q1.append(q1)
attend_stroketeam_mean.append(data_stroke_team.mean())
attend_stroketeam_q3.append(q3)
attend_stroketeam_max.append(data_stroke_team.max())
# Create dataframe with six columns (hospital and descriptive stats)
df = pd.DataFrame(hospital_names, columns=[f"hospital_{prefix_text}"])
df[f"shap_min_{prefix_text}"] = attend_stroketeam_min
df[f"shap_q1_{prefix_text}"] = attend_stroketeam_q1
df[f"shap_mean_{prefix_text}"] = attend_stroketeam_mean
df[f"shap_q3_{prefix_text}"] = attend_stroketeam_q3
df[f"shap_max_{prefix_text}"] = attend_stroketeam_max
return(df)
Calculate the descriptive stats (IQR values) for the range of SHAP values (and main effect) for the attended hospital.
df_me = shap_descriptive_stats(hospital_names, df_hosp_shap_main_effects, "me")
df_sv = shap_descriptive_stats(hospital_names, df_hosp_shap_values, "sv")
Join the dataframes together (SHAP valeus and SHAP main effect) in one dataframe
df_hosp_shap_descriptive_stats = df_me.join(df_sv)
df_hosp_shap_descriptive_stats.drop(columns=["hospital_sv"], inplace=True)
df_hosp_shap_descriptive_stats.rename(columns={"hospital_me": "hospital"},
inplace=True)
Sort in descending SHAP main effect value order
df_hosp_shap_descriptive_stats.sort_values("shap_mean_me",
ascending=False, inplace=True)
df_hosp_shap_descriptive_stats.head(5)
| hospital | shap_min_me | shap_q1_me | shap_mean_me | shap_q3_me | shap_max_me | shap_min_sv | shap_q1_sv | shap_mean_sv | shap_q3_sv | shap_max_sv | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | CNBGF2713O | 1.195877 | 1.314107 | 1.351095 | 1.393786 | 1.508929 | 0.648626 | 1.160532 | 1.338743 | 1.513107 | 1.989456 |
| 25 | GKONI0110I | 0.767646 | 1.057928 | 1.110841 | 1.168451 | 1.358355 | 0.046286 | 0.888337 | 1.085008 | 1.286739 | 1.779779 |
| 65 | NTPQZ0829K | 0.797407 | 0.978037 | 1.006841 | 1.050939 | 1.206905 | 0.209000 | 0.763998 | 0.917956 | 1.126985 | 1.599991 |
| 62 | MHMYL4920B | 0.753430 | 0.946048 | 0.964973 | 0.986911 | 1.103224 | 0.130139 | 0.786147 | 0.933300 | 1.059232 | 1.476519 |
| 32 | HPWIF9956L | 0.750452 | 0.902049 | 0.956728 | 1.008806 | 1.169653 | 0.206888 | 0.652731 | 0.901027 | 1.152177 | 1.624375 |
Want to add admission figures to xlabel in boxplot. Create dataframe with admissions and thrombolysis rate per stroke team (index)
# Get list of unique stroke team names
unique_stroketeams_list = list(set(X_combined["Stroke team"]))
# Calculate admissions to each team
admissions = [X[f'team_{s}'].sum() for s in unique_stroketeams_list]
# Create dataframe with stroke team and admissions
df_stroketeam_ivt_adms = pd.DataFrame(unique_stroketeams_list,
columns=["Stroke team"])
df_stroketeam_ivt_adms["Admissions"] = admissions
df_stroketeam_ivt_adms.set_index("Stroke team", inplace=True)
df_stroketeam_ivt_adms.sort_values("Admissions", ascending=True, inplace=True)
# Calculate IVT rate per hosptial
hosp_ivt_rate = data.groupby(by=["Stroke team"]).mean()["Thrombolysis"]
# Join IVT rate with admissions per hosptial
df_stroketeam_ivt_adms = df_stroketeam_ivt_adms.join(hosp_ivt_rate)
df_stroketeam_ivt_adms
| Admissions | Thrombolysis | |
|---|---|---|
| Stroke team | ||
| JXJYG0100P | 120 | 0.233333 |
| VVDIY0129H | 130 | 0.192308 |
| YEXCH8391J | 149 | 0.228188 |
| CNBGF2713O | 152 | 0.480263 |
| XPABC1435F | 166 | 0.216867 |
| ... | ... | ... |
| JINXD0311F | 1377 | 0.368192 |
| AKCGO9726K | 1428 | 0.369748 |
| OFKDF3720W | 1488 | 0.228495 |
| FPTBM7594G | 1645 | 0.321581 |
| FAJKD7118X | 1980 | 0.275758 |
132 rows × 2 columns
Create data for boxplot
# Go through order of hospital in Dataframe (ordered on mean SHAP main effect)
hospital_order = df_hosp_shap_descriptive_stats["hospital"]
# Initiate lists
# 1) SHAP main effect (one per hospital) for instances that attend stroke team
# 2) SHAP value (one per hospital) for instances that attend stroke team
me_attend_stroketeam_groups_ordered = []
sv_attend_stroketeam_groups_ordered = []
# Initiate lists
# 1) SHAP main effect (one per hospital) for instances that don't attend stroke
# team
# 2) SHAP value (one per hospital) for instances that don't attend stroke team
me_not_attend_stroketeam_groups_ordered = []
sv_not_attend_stroketeam_groups_ordered = []
# Initiate list for xlabels for boxplot "stroke team name (admissions)"
xlabel = []
# Through hospital in defined order (as determined above)
for h in hospital_order:
# Attend
mask = df_hosp_shap_main_effects['Stroke team'] == h
me_attend_stroketeam_groups_ordered.append(
df_hosp_shap_main_effects[h][mask])
sv_attend_stroketeam_groups_ordered.append(df_hosp_shap_values[h][mask])
# Not attend
mask = df_hosp_shap_main_effects['Stroke team'] != h
me_not_attend_stroketeam_groups_ordered.append(
df_hosp_shap_main_effects[h][mask])
sv_not_attend_stroketeam_groups_ordered.append(df_hosp_shap_values[h][mask])
# Label
ivt_rate = int(df_stroketeam_ivt_adms['Thrombolysis'].loc[h] * 100)
xlabel.append(f"{h} ({df_stroketeam_ivt_adms['Admissions'].loc[h]}, "
f"{ivt_rate}%)")
Resource for using overall y min and max of both datasets on the 4 plots so have the same range https://blog.finxter.com/how-to-find-the-minimum-of-a-list-of-lists-in-python/#:~:text=With%20the%20key%20argument%20of,of%20the%20list%20of%20lists.
# Plot 34 hospitals on each figure to aid visually
print("Shows the range of contributions to the prediction from this hospital "
"when patients attend this hosptial")
# To group the hospitals into 34
st = 0
ed = 34
inc = ed
max_size = n_hospitals
# Use overall y min & max of both datasets on the 4 plots so have same range
ymin = min(min(sv_attend_stroketeam_groups_ordered, key=min))
ymax = max(max(sv_attend_stroketeam_groups_ordered, key=max))
# Adjust min and max to accommodate some wriggle room
yrange = ymax - ymin
ymin = ymin - yrange/50
ymax = ymax + yrange/50
# Create figure
fig = plt.figure(figsize=(15,25))
# Create four subplots (divide the 132 hospitals across these to ai visability)
for subplot in range(4):
ax = fig.add_subplot(4,1,subplot+1)
# The contribution from this hospital when patients do not attend this
# hospital
ticks = xlabel[st:ed]
pos_sv = np.array(range(
len(sv_attend_stroketeam_groups_ordered[st:ed])))*2.0-0.4
bp_sv = plt.boxplot(sv_attend_stroketeam_groups_ordered[st:ed],
positions=pos_sv, sym='', whis=99999, widths=0.6)
pos_me = np.array(range(len(
me_attend_stroketeam_groups_ordered[st:ed])))*2.0+0.4
bp_me = plt.boxplot(me_attend_stroketeam_groups_ordered[st:ed],
positions=pos_me, sym='', whis=99999, widths=0.6)
# colors are from http://colorbrewer2.org/
set_box_color(bp_me, '#D7191C')
set_box_color(bp_sv, '#2C7BB6')
# draw temporary red and blue lines and use them to create a legend
plt.plot([], c='#2C7BB6', label='SHAP value')
plt.plot([], c='#D7191C', label='SHAP main effect')
plt.legend()
plt.xticks(range(0, len(ticks) * 2, 2), ticks)
plt.xlim(-2, len(ticks)*2)
plt.ylim(ymin, ymax)
plt.tight_layout()
# Add line at Shap = 0
plt.plot([plt.xlim()[0], plt.xlim()[1]], [0,0], c='0.8')
plt.ylabel('SHAP (value or main effect) of one-hot encoded feature',
size=12)
plt.xlabel('Stroke team (admissions, IVT rate %)', size=12)
plt.xticks(rotation=90)
st = min(st+inc,max_size)
ed = min(ed+inc,max_size)
plt.subplots_adjust(bottom=0.25, wspace=0.05)
plt.tight_layout(pad=2)
plt.savefig(f'./output/{notebook}_{model_text}_individual_hosp_shap_value_and_'
f'main_effect_attend_vs_notattend_boxplot.jpg', dpi=300,
bbox_inches='tight', pad_inches=0.2)
plt.show()
Shows the range of contributions to the prediction from this hospital when patients attend this hosptial
Count number of hospitals with IQR below, spanning, on or above zero#
Notice that when patients do not attend the hospital the range of the SHAP values are largely centred on zero. When patients do attend hosptial, the range of SHAP values are largely one side of zero or the other (only a minority of hospitals have their interquartile range spanning zero).
def count_hospitals_in_(q1, q3):
"""
Given the upper and lower interquartile values (each in a series), return
the number of times the interquartile range is totally below zero, spans
zero, above zero, is zero.
q1 [Series]: lower interquartile range
q3 [Series]: upper interquartile range
return [(integer, integer, integer, integer)]
"""
n_iqr_below_zero = q3 < 0
n_iqr_spans_zero = q1 * q3
n_iqr_above_zero = q1 > 0
n_iqr_is_zero1 = q1 == 0
n_iqr_is_zero2 = q3 == 0
n_iqr_is_zero = n_iqr_is_zero1 * n_iqr_is_zero2
return(n_iqr_below_zero, n_iqr_spans_zero, n_iqr_above_zero, n_iqr_is_zero)
(n_iqr_below_zero_me, n_iqr_spans_zero_me,
n_iqr_above_zero_me, n_iqr_is_zero_me) = (
count_hospitals_in_(df_hosp_shap_descriptive_stats["shap_q1_me"],
df_hosp_shap_descriptive_stats["shap_q3_me"]))
(n_iqr_below_zero_sv, n_iqr_spans_zero_sv,
n_iqr_above_zero_sv, n_iqr_is_zero_sv) = (
count_hospitals_in_(df_hosp_shap_descriptive_stats["shap_q1_sv"],
df_hosp_shap_descriptive_stats["shap_q3_sv"]))
print (f"Number hospitals whose interquartile range is below zero. "
f"Main effect: {n_iqr_below_zero_me.sum()}."
f" Shap values: {n_iqr_below_zero_sv.sum()}")
print (f"Number hospitals whose interquartile range spans zero. "
f"Main effect: {n_iqr_spans_zero_me.lt(0).sum()}. "
f"Shap values: {n_iqr_spans_zero_sv.lt(0).sum()}")
print (f"Number hospitals whose interquartile range is above zero. "
f"Main effect: {n_iqr_above_zero_me.sum()}. "
f"Shap values: {n_iqr_above_zero_sv.sum()}")
print (f"Number hospitals whose interquartile range is zero. "
f"Main effect: {n_iqr_is_zero_me.sum()}. "
f"Shap values: {n_iqr_is_zero_sv.sum()}")
Number hospitals whose interquartile range is below zero. Main effect: 68. Shap values: 58
Number hospitals whose interquartile range spans zero. Main effect: 2. Shap values: 24
Number hospitals whose interquartile range is above zero. Main effect: 58. Shap values: 46
Number hospitals whose interquartile range is zero. Main effect: 4. Shap values: 4
How does the SHAP value (and SHAP main effect) for the one-hot encoded hospital features compare to the thrombolysis rate of the hospital?#
Create dataframe containing the hospital’s IVT rate and the attended hosptial’s SHAP value and SHAP main effect.
# Calculate IVT rate per hosptial
hosp_ivt_rate = data.groupby(by=["Stroke team"]).mean()["Thrombolysis"]
# Join IVT rate with mean SHAP value per hosptial
df_hosp_plot = (
df_hosp_shap_descriptive_stats[["shap_mean_sv","hospital"]].copy(deep=True))
df_hosp_plot.set_index("hospital", inplace=True)
df_hosp_plot = df_hosp_plot.join(hosp_ivt_rate)
# Join IVT rate with mean SHAP main effect per hosptial
temp = (
df_hosp_shap_descriptive_stats[["shap_mean_me","hospital"]].copy(deep=True))
temp.set_index("hospital", inplace=True)
df_hosp_plot = df_hosp_plot.join(temp)
Save dataframe as csv file.
filename = (f'./output/{notebook}_{model_text}_hospital_shap_vs_ivt_rate.csv')
df_hosp_plot.to_csv(filename)
Plot SHAP value for one-hot encoded hospital feature (mean SHAP value and mean SHPA main effect for those instances that attend the hospital) vs hospital IVT rate
# Setup data for chart
x1 = df_hosp_plot['shap_mean_me']
x2 = df_hosp_plot['shap_mean_sv']
y = df_hosp_plot['Thrombolysis']
# Fit a regression line to the x1 points
slope1, intercept1, r_value1, p_value1, std_err1 = \
stats.linregress(x1, y)
r_square1 = r_value1 ** 2
y_pred1 = intercept1 + (x1 * slope1)
# Fit a regression line to the x2 points
slope2, intercept2, r_value2, p_value2, std_err2 = \
stats.linregress(x2, y)
r_square2 = r_value2 ** 2
y_pred2 = intercept2 + (x2 * slope2)
# Create scatter plot with regression line
fig = plt.figure(figsize=(8,8))
ax = fig.add_subplot(1,1,1)
ax.scatter(x1, y, color = "blue", marker="+", s=30)
ax.scatter(x2, y, color = "orange", marker="x", s=20)
ax.plot (x1, y_pred1, color = 'blue', linestyle=':')
f1 = ('formula: ' + str("{:.2f}".format(slope1)) + 'x + ' +
str("{:.2f}".format(intercept1)))
text1 = (f'SHAP main effect results (blue)\nR squared: {r_square1:.3f}\np: '
f'{p_value1:0.4f}\nformula: {f1}')
ax.text(-1.3, 0.45, text1,
bbox=dict(facecolor='white', edgecolor='black'))
ax.plot (x2, y_pred2, color = 'orange', linestyle=':')
f2 = ('formula: ' + str("{:.2f}".format(slope2)) + 'x + ' +
str("{:.2f}".format(intercept2)))
text2 = (f'SHAP value results (orange)\nR squared: {r_square2:.3f}\np: '
f'{p_value2:0.4f}\nformula: {f2}')
ax.text(-1.3, 0.39, text2,
bbox=dict(facecolor='white', edgecolor='black'))
ax.set_xlabel("SHAP (value or main effect) "
"[mean of the instances that attend the hospital]")
ax.set_ylabel('Thrombolysis rate')
ax.set_title("SHAP (value or main effect) for one-hot encoded hospital feature "
"vs IVT rate")
plt.grid()
plt.savefig(f'./output/{notebook}_{model_text}'
f'_attended_hosp_shap_value_and_main_effect_vs_ivt_rate.jpg',
dpi=300, bbox_inches='tight', pad_inches=0.2)
plt.show()