Distance-based treatment effects (plot for different stroke types)#
Based on the figures from Holodinsky et al. 2017 - “Drip and Ship Versus Direct to Comprehensive Stroke Center”
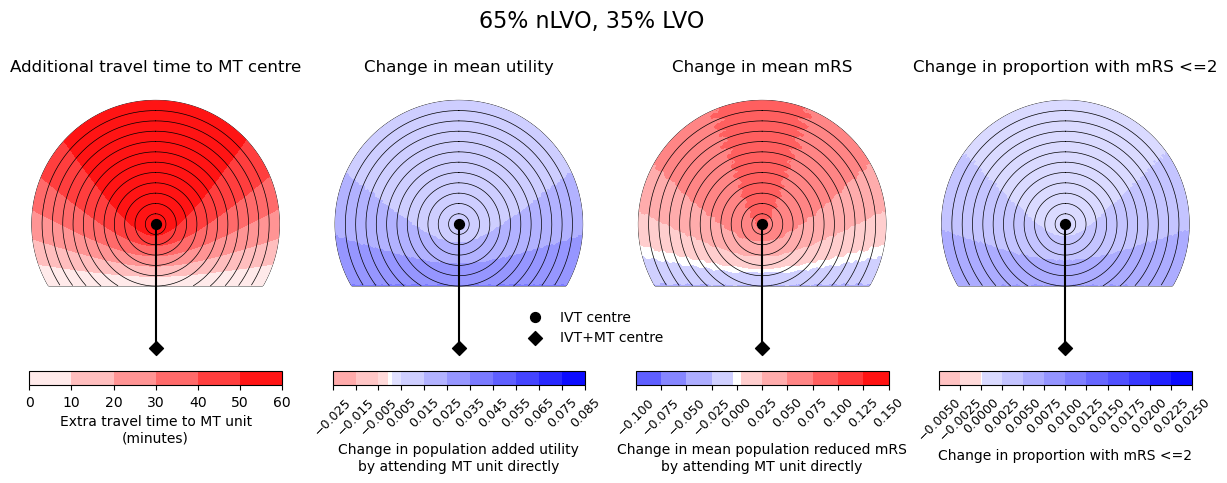
Here we show the difference in outcome measures for nLVO, LVO, and a mixed population (65% nLVO, 35% LVO). There is 60 min travel time between IVT and MT centres.
Notebook admin#
# Import packages
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Keep notebook cleaner once finalised
import warnings
warnings.filterwarnings('ignore')
Assumptions#
Fixed times for additional delays:
# All fixed times have units of minutes
fixed_times = dict(
onset_to_ambulance_arrival = 60,
ivt_arrival_to_treatment = 30,
transfer_additional_delay = 60,
travel_ivt_to_mt = 50,
mt_arrival_to_treatment = 90,
)
Define a helper function to build the time grid:
def make_time_grid(xy_max, step, x_offset=0, y_offset=0):
# Times for each row....
x_times = np.arange(-xy_max, xy_max + step, step) - x_offset
# ... and each column.
y_times = np.arange(-xy_max, xy_max + step, step) - y_offset
# The offsets shift the position of (0,0) from the grid centre
# to (x_offset, y_offset). Distances will be calculated from the
# latter point.
# Mesh to create new grids by stacking rows (xx) and columns (yy):
xx, yy = np.meshgrid(x_times, y_times)
# Then combine the two temporary grids to find distances:
radial_times = np.sqrt(xx**2.0 + yy**2.0)
return radial_times
Build the grids:
def make_grids_travel_time(time_travel_ivt_to_mt, time_travel_max, ivt_coords, mt_coords, grid_step=1):
# Make the grid a bit larger than the max travel time:
grid_xy_max = time_travel_max + grid_step*2
grid_time_travel_directly_to_ivt = make_time_grid(
grid_xy_max, grid_step, x_offset=ivt_coords[0],
y_offset=ivt_coords[1])
grid_time_travel_directly_to_mt = make_time_grid(
grid_xy_max, grid_step, x_offset=mt_coords[0], y_offset=mt_coords[1])
grid_time_travel_directly_diff = (
grid_time_travel_directly_to_ivt - grid_time_travel_directly_to_mt)
extent = [-grid_xy_max - grid_step*0.5,
+grid_xy_max - grid_step*0.5,
-grid_xy_max - grid_step*0.5,
+grid_xy_max - grid_step*0.5]
return (grid_time_travel_directly_to_ivt,
grid_time_travel_directly_to_mt,
grid_time_travel_directly_diff,
extent)
Plot travel times#
Grids#
from outcome_utilities.geography_plot import find_mask_within_flattened_circle
Update plotting style:
time_step_circle = 5
circ_linewidth = 0.5
from outcome_utilities.geography_plot import circle_plot
def make_grids_treatment_time(
grid_time_travel_directly_to_ivt,
grid_time_travel_directly_to_mt,
times_dir):
"""
Define new treatment time grids.
"""
grid_time_ivt_at_ivtcentre = (
times_dir['onset_to_ambulance_arrival'] +
grid_time_travel_directly_to_ivt +
times_dir['ivt_arrival_to_treatment']
)
grid_time_ivt_at_ivt_then_mt_at_mtcentre = (
times_dir['onset_to_ambulance_arrival'] +
grid_time_travel_directly_to_ivt +
times_dir['ivt_arrival_to_treatment'] +
times_dir['transfer_additional_delay'] +
times_dir['travel_ivt_to_mt'] +
times_dir['mt_arrival_to_treatment']
)
grid_time_ivt_at_mtcentre = (
times_dir['onset_to_ambulance_arrival'] +
grid_time_travel_directly_to_mt +
times_dir['ivt_arrival_to_treatment']
)
grid_time_ivt_then_mt_at_mtcentre = (
times_dir['onset_to_ambulance_arrival'] +
grid_time_travel_directly_to_mt +
times_dir['ivt_arrival_to_treatment'] +
times_dir['mt_arrival_to_treatment']
)
return (grid_time_ivt_at_ivtcentre,
grid_time_ivt_at_ivt_then_mt_at_mtcentre,
grid_time_ivt_at_mtcentre,
grid_time_ivt_then_mt_at_mtcentre)
from outcome_utilities.clinical_outcome import Clinical_outcome
mrs_dists = pd.read_csv(
'./outcome_utilities/mrs_dist_probs_cumsum.csv', index_col='Stroke type')
# Set up outcome model
outcome_model = Clinical_outcome(mrs_dists)
def find_grid_outcomes(outcome_model, grid_time_ivt, grid_time_mt,
patient_props):
"""
For all pairs of treatment times, calculate the changes in utility
and mRS for the patient population.
Inputs:
Returns:
"""
grid_shape = grid_time_ivt.shape
utility_grid = np.empty(grid_shape)
mRS_grid = np.empty(grid_shape)
good_outcomes_grid = np.empty(grid_shape)
for row in range(grid_shape[0]):
for col in range(int(grid_shape[1]*0.5)+1):
# ^ weird range is for symmetry later when filling grids.
# Expect col_opp, the opposite column as reflected in the
# x=0 axis, to contain the same values as col:
col_opp = grid_shape[1]-1-col
time_to_ivt = grid_time_ivt[row, col]
time_to_mt = grid_time_mt[row, col]
outcomes = outcome_model.calculate_outcomes(
time_to_ivt, time_to_mt, patients=1000)
# Find the change in utility:
added_utility = find_weighted_change(
outcomes['lvo_ivt_added_utility'],
outcomes['lvo_mt_added_utility'],
outcomes['nlvo_ivt_added_utility'],
patient_props
)
# Add this value to the grid:
utility_grid[row,col] = added_utility
utility_grid[row,col_opp] = added_utility
# Find the change in mRS:
reduced_mRS = find_weighted_change(
outcomes['lvo_ivt_mean_shift'],
outcomes['lvo_mt_mean_shift'],
outcomes['nlvo_ivt_mean_shift'],
patient_props
)
# Add this value to the grid:
mRS_grid[row,col] = reduced_mRS
mRS_grid[row,col_opp] = reduced_mRS
# Find change in proportion with mRS <=2
nlvo_ivt_added_good_outcomes = (
outcomes['nlvo_ivt_cum_probs'][2] -
outcomes['nlvo_untreated_cum_probs'][2])
lvo_ivt_added_good_outcomes = (
outcomes['lvo_ivt_cum_probs'][2] -
outcomes['lvo_untreated_cum_probs'][2])
lvo_mt_added_good_outcomes = (
outcomes['lvo_mt_cum_probs'][2] -
outcomes['lvo_untreated_cum_probs'][2])
added_good_outcomes = find_weighted_change(
lvo_ivt_added_good_outcomes,
lvo_mt_added_good_outcomes,
nlvo_ivt_added_good_outcomes,
patient_props
)
good_outcomes_grid[row,col] = added_good_outcomes
good_outcomes_grid[row,col_opp] = added_good_outcomes
# Adjust outcome for just treated population
utility_grid = utility_grid / patient_props['treated_population']
mRS_grid = mRS_grid / patient_props['treated_population']
return utility_grid, mRS_grid, good_outcomes_grid
def find_weighted_change(change_lvo_ivt, change_lvo_mt, change_nlvo_ivt,
patient_props):
"""
Take the total changes for each category and calculate their
weighted sum, where weights are from the proportions of the
patient population.
(originally from matrix notebook)
Inputs:
Returns:
"""
# If LVO-IVT is greater change than LVO-MT then adjust MT for
# proportion of patients receiving IVT:
if change_lvo_ivt > change_lvo_mt:
diff = change_lvo_ivt - change_lvo_mt
change_lvo_mt += diff * patient_props['lvo_mt_also_receiving_ivt']
# Calculate changes multiplied by proportions (cp):
cp_lvo_mt = (
change_lvo_mt *
patient_props['lvo'] *
patient_props['lvo_treated_ivt_mt']
)
cp_lvo_ivt = (
change_lvo_ivt *
patient_props['lvo'] *
patient_props['lvo_treated_ivt_only']
)
cp_nlvo_ivt = (
change_nlvo_ivt *
patient_props['nlvo'] *
patient_props['nlvo_treated_ivt_only']
)
total_change = cp_lvo_mt + cp_lvo_ivt + cp_nlvo_ivt
return total_change
Combined circle plot#
Share these colour limits between all figures:
vmin_time = 0
vmax_time = 60
vmin_util = -0.025 # -0.045
vmax_util = 0.08 # 0.035
vmin_mRS = -0.1 # -0.250
vmax_mRS = 0.15 # 0.175
vmin_good = -0.005
vmax_good = 0.025
Make new colour maps that are based on a diverging colour map but have the zero (white) off-centre:
from outcome_utilities.geography_plot import make_new_cmap_diverging
cmap_util = make_new_cmap_diverging(
vmin_util, vmax_util, cmap_base='bwr_r', cmap_name='bwr_util')
cmap_mRS = make_new_cmap_diverging(
vmin_mRS, vmax_mRS, cmap_base='bwr', cmap_name='bwr_mRS')
cmap_good = make_new_cmap_diverging(
vmin_good, vmax_good, cmap_base='bwr_r', cmap_name='bwr_good')
# For the time colours, take only the red half of 'bwr':
from matplotlib import colors
colours_time = plt.get_cmap('bwr')(np.linspace(0.5, 1.0, 256))
cmap_time = colors.LinearSegmentedColormap.from_list('bwr_time', colours_time)
Define contour levels for plotting:
from outcome_utilities.geography_plot import make_levels_with_zeroish
level_step_util = 0.01 # 0.005
levels_util = make_levels_with_zeroish(
level_step_util, vmax_util, zeroish=1e-3, vmin=vmin_util)
level_step_mRS = 0.025 # 0.025
levels_mRS = make_levels_with_zeroish(
level_step_mRS, vmax_mRS, zeroish=4e-3, vmin=vmin_mRS)
level_step_good_outcome = 0.0025 # 0.025
levels_good_outcome = make_levels_with_zeroish(
level_step_good_outcome, vmax_good, zeroish=5e-5, vmin=vmin_good)
The following big cell calculates the new grids and then makes the plots.
time_travel_ivt_to_mt = 60
time_onset_to_ambulance_arrival = 60
patient_props_list = [
dict(
lvo = 0,
nlvo = 1, # 1-LVO
lvo_mt_also_receiving_ivt = 0.85,
lvo_treated_ivt_only = 0.0,
lvo_treated_ivt_mt = 0.286, # 0.286 gives 10% final MT if 35%LVO
nlvo_treated_ivt_only = 0.155, # 0.155 gives final 20% IVT
),
dict(
lvo = 1,
nlvo = 0, # 1-LVO
lvo_mt_also_receiving_ivt = 0.85,
lvo_treated_ivt_only = 0.0,
lvo_treated_ivt_mt = 0.286, # 0.286 gives 10% final MT if 35%LVO
nlvo_treated_ivt_only = 0.155, # 0.155 gives final 20% IVT
),
dict(
lvo = 0.35,
nlvo = 1.0-0.35, # 1-LVO
lvo_mt_also_receiving_ivt = 0.85,
lvo_treated_ivt_only = 0.0,
lvo_treated_ivt_mt = 0.286, # 0.286 gives 10% final MT if 35%LVO
nlvo_treated_ivt_only = 0.155, # 0.155 gives final 20% IVT
)
]
#patient_props_list = [patient_props_list[0]]
titles = ['nLVO', 'LVO', '65% nLVO, 35% LVO']
# Recalculate some values:
ivt_coords = [0, 0]
mt_coords = [0, -time_travel_ivt_to_mt]
# Only calculate travel times up to this x or y displacement:
time_travel_max = time_travel_ivt_to_mt
# Change how granular the grid is.
grid_step = 1 # minutes
# Make the travel time grids:
(grid_time_travel_directly_to_ivt,
grid_time_travel_directly_to_mt,
grid_time_travel_directly_diff,
extent) = \
make_grids_travel_time(time_travel_ivt_to_mt, time_travel_max,
ivt_coords, mt_coords, grid_step=1)
# Find grid coordinates within the largest circle:
grid_mask = find_mask_within_flattened_circle(
grid_time_travel_directly_diff,
grid_time_travel_directly_to_ivt,
time_travel_max)
coords_valid = np.where(grid_mask<1)
for count, patient_props in enumerate(patient_props_list):
treated_population = (
patient_props['nlvo'] * patient_props['nlvo_treated_ivt_only'] +
patient_props['lvo'] * patient_props['lvo_treated_ivt_mt'] +
patient_props['lvo'] * patient_props['lvo_treated_ivt_only']
)
patient_props['treated_population'] = treated_population
# Build a new dictionary of fixed times:
times_dir = {}
for key in fixed_times:
times_dir[key] = fixed_times[key]
times_dir['onset_to_ambulance_arrival'] = time_onset_to_ambulance_arrival
times_dir['travel_ivt_to_mt'] = time_travel_ivt_to_mt
# Grids of times to treatment:
(grid_time_ivt_at_ivtcentre,
grid_time_ivt_at_ivt_then_mt_at_mtcentre,
grid_time_ivt_at_mtcentre,
grid_time_ivt_then_mt_at_mtcentre) = \
make_grids_treatment_time(grid_time_travel_directly_to_ivt,
grid_time_travel_directly_to_mt, times_dir)
# Grids of changed outcomes
# Case 1: IVT at the IVT centre, then MT at the IVT/MT centre
grid_utility_case1, grid_mRS_case1, grid_good_outcomes_case1 = find_grid_outcomes(
outcome_model,
grid_time_ivt_at_ivtcentre,
grid_time_ivt_at_ivt_then_mt_at_mtcentre,
patient_props
)
# Case 2: IVT at the IVT/MT centre, then MT at IVT/MT centre
grid_utility_case2, grid_mRS_case2, grid_good_outcomes_case2 = find_grid_outcomes(
outcome_model,
grid_time_ivt_at_mtcentre,
grid_time_ivt_then_mt_at_mtcentre,
patient_props
)
# Difference between them:
grid_utility_diff = grid_utility_case2 - grid_utility_case1
grid_mRS_diff = grid_mRS_case2 - grid_mRS_case1
grid_good_outcomes_diff = grid_good_outcomes_case2 - grid_good_outcomes_case1
print('-'*50)
print(f'Time of travel from IVT to MT: {time_travel_ivt_to_mt:2d}')
print(f'Time from onset to ambulance arrival: ',
f'{time_onset_to_ambulance_arrival:2d}')
print(f'Utility {np.min(grid_utility_diff[coords_valid]):7.4f} '+
f'{np.max(grid_utility_diff[coords_valid]):7.4f}')
print(f'mRS {np.min(grid_mRS_diff[coords_valid]):7.4f} '+
f'{np.max(grid_mRS_diff[coords_valid]):7.4f}')
print(f'Good outcomes {np.min(grid_good_outcomes_diff[coords_valid]):7.4f} '+
f'{np.max(grid_good_outcomes_diff[coords_valid]):7.4f}')
# Plot setup:
fig, axs = plt.subplots(2, 4, figsize=(15,4),
gridspec_kw={'wspace':0.2, 'hspace':0.0, 'height_ratios':[20,1]})
ax_time = axs[0,0]
ax_time_cbar = axs[1,0]
ax_util = axs[0,1]
ax_util_cbar = axs[1,1]
ax_mRS = axs[0,2]
ax_mRS_cbar = axs[1,2]
ax_good = axs[0,3]
ax_good_cbar = axs[1,3]
# Travel time:
circle_plot(
-grid_time_travel_directly_diff,
time_travel_ivt_to_mt,
time_travel_max,
time_step_circle,
vmin_time,
vmax_time,
ivt_coords=ivt_coords,
mt_coords=mt_coords,
extent=extent,
imshow=0,
ax=ax_time,
cax=ax_time_cbar,
cmap=cmap_time,
cbar_label='Extra travel time to MT unit\n(minutes)',
cbar_orientation='horizontal',
n_contour_steps=6)
ax_time.set_title('Additional travel time to MT centre')
# Draw legend now:
fig.legend(loc='upper center', bbox_to_anchor=[0.5,0.33], ncol=1, frameon=False)
# Good outcomes
circle_plot(
grid_good_outcomes_diff,
time_travel_ivt_to_mt,
time_travel_max,
time_step_circle,
vmin_good,
vmax_good,
ivt_coords=ivt_coords,
mt_coords=mt_coords,
extent=extent,
imshow=0,
ax=ax_good,
cax=ax_good_cbar,
cmap=cmap_good,
cbar_label='Change in proportion with mRS <=2',
cbar_orientation='horizontal',
# cbar_format_str='{:3.2f}',
levels=levels_good_outcome,
cbar_ticks=np.arange(
vmin_good, vmax_good+level_step_good_outcome, level_step_good_outcome))
ax_good.set_title('Change in proportion with mRS <=2')
# mRS:
circle_plot(
grid_mRS_diff,
time_travel_ivt_to_mt,
time_travel_max,
time_step_circle,
vmin_mRS,
vmax_mRS,
ivt_coords=ivt_coords,
mt_coords=mt_coords,
extent=extent,
imshow=0,
ax=ax_mRS,
cax=ax_mRS_cbar,
cmap=cmap_mRS,
cbar_label='Change in mean population reduced mRS\n'+
'by attending MT unit directly',
cbar_orientation='horizontal',
# cbar_format_str='{:3.2f}',
levels=levels_mRS,
cbar_ticks=np.arange(
vmin_mRS, vmax_mRS+level_step_mRS, level_step_mRS))
ax_mRS.set_title('Change in mean mRS')
# Utility:
circle_plot(
grid_utility_diff,
time_travel_ivt_to_mt,
time_travel_max,
time_step_circle,
vmin_util,
vmax_util,
ivt_coords=ivt_coords,
mt_coords=mt_coords,
extent=extent,
imshow=0,
ax=ax_util,
cax=ax_util_cbar,
cmap=cmap_util,
cbar_label=('Change in population added utility\n'+
'by attending MT unit directly'),
cbar_orientation='horizontal',
levels=levels_util,
# cbar_format_str='{:3.3f}',
cbar_ticks=np.arange(
vmin_util, vmax_util+level_step_util, level_step_util))
ax_util.set_title('Change in mean utility')
for ax in [ax_time, ax_util, ax_mRS, ax_good]:
ax.set_xlabel('')
ax.set_ylabel('')
ax.set_xticks([])
ax.set_yticks([])
for spine in ['top', 'bottom', 'left', 'right']:
ax.spines[spine].set_color('None')
ax.set_ylim(-70,70)
# Reduce font size of ax.xaxis.label
ax.tick_params(axis='both', which='major', labelsize=9)
# Rotate colourbar tick labels to prevent overlapping:
for ax in [ax_mRS_cbar, ax_util_cbar, ax_good_cbar]:
for tick in ax.get_xticklabels():
tick.set_rotation(45)
tick.set_fontsize(9)
tick.set_horizontalalignment('center')
# Set supertitle
fig.suptitle(titles[count], fontsize=16, y=1.05)
filename = (f'circle_plots_stroke_type_{titles[count]}_60_60')
plt.savefig('./images/'+filename+'.jpg', dpi=300, bbox_inches='tight')
plt.show()
--------------------------------------------------
Time of travel from IVT to MT: 60
Time from onset to ambulance arrival: 60
Utility -0.0239 0.0000
mRS 0.0000 0.1360
Good outcomes -0.0040 0.0000
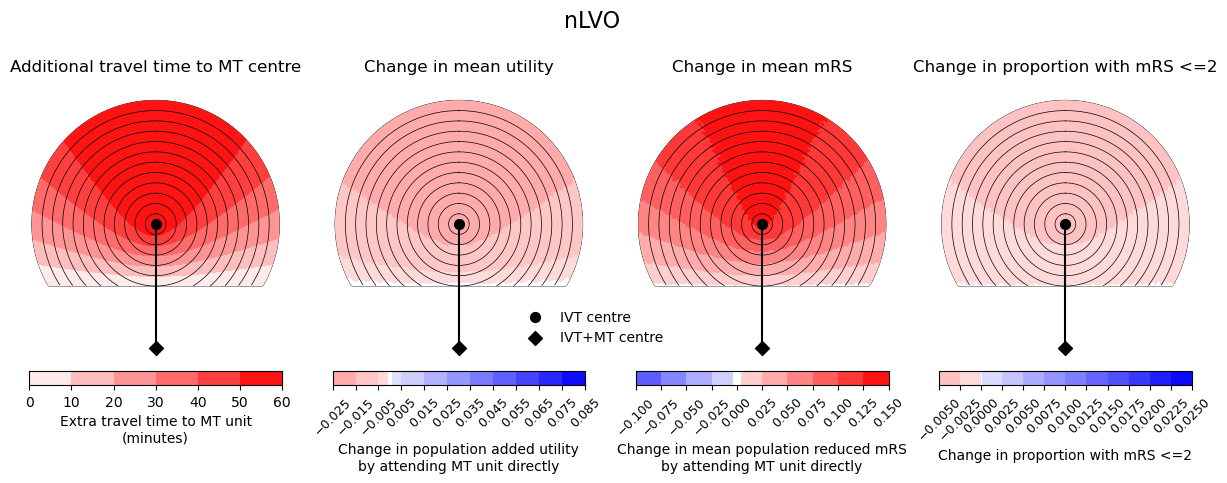
--------------------------------------------------
Time of travel from IVT to MT: 60
Time from onset to ambulance arrival: 60
Utility 0.0345 0.0721
mRS -0.0569 0.0255
Good outcomes 0.0103 0.0217
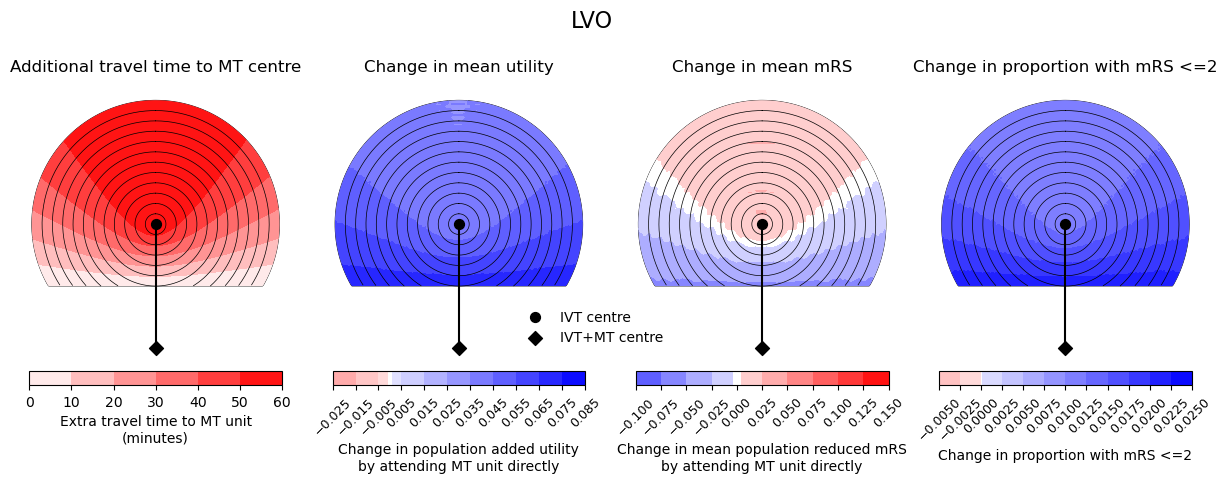
--------------------------------------------------
Time of travel from IVT to MT: 60
Time from onset to ambulance arrival: 60
Utility 0.0054 0.0360
mRS -0.0283 0.0800
Good outcomes 0.0010 0.0076