Calculating outcome probabilities with time to treatment#
In this notebook we explain the simplest way to find probability and odds depending on time to treatment that will be used in predicting modified Rankin Scale (mRS) after stroke.
Plain English summary#
We have previously derived the probabilities of mRS at time zero, \(t=0\,\)hr, and the time of no effect, \(t=t_{\mathrm{ne}}\). We need to use the data we have at these two fixed times to create a model that describes probability at any time \(t\).
Since we know that probability, odds, and log(odds) are all linked, TO DO: link to some intro notebook we can find the outcomes for just one of these measures and then convert the outcomes into any of the other measures.
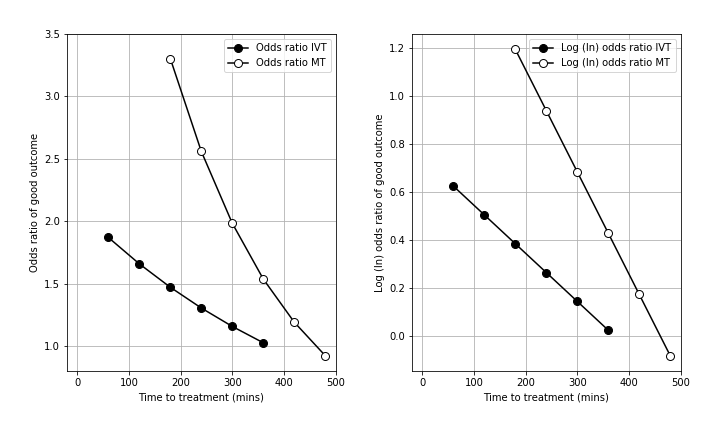
We’ll find the outcomes for log(odds) because these are the easiest to calculate. In the literature, the log(odds ratio) has been presented at regular time intervals and falls off approximately linearly with time \(t\). This allows it to be modelled as a straight line.
The right-hand plot shows the falling log-odds ratio with time. It uses data for thrombolysis from Emberson et al. 2014 and data for thrombectomy from Fransen et al. 2016.
We’ll then convert the outcomes from log(odds) into probability.
Method#
The steps here are, for mRS\(\leq\)5\(^*\):
Define log(odds) with time as a straight line.
Find the proportion of time to onset to time of no effect that matches the time to treatment.
Take the same proportion from the log-odds at time of onset to the log-odds at time of no effect. This is the log-odds at time of treatment.
Convert the log-odds to probability.
Repeat this for the other mRS bands to find the mRS distribution at the time of treatment.
\(^*\) We cannot include mRS\(\leq\)6 in these calculations, but luckily we already know that \(P(\mathrm{mRS}\leq6)=1.0\) at all \(t\).
Note: in this document we calculate log(odds) instead of log(odds radio) like in the plot above. It doesn’t make any difference to the final result. The log(odds ratio) is used so that it is clear at what time the log(odds ratio) falls below zero, and so when to define the time of no effect. Since we are using data where we already know this time of no effect and the probability distributions at that time, we can skip a step and directly model log(odds) as a straight line. We can make this simplification because the straight line fits to log(odds) and to log(odds ratio) share a gradient and only differ in their start and end values (their displacement in \(y\)).
Notebook admin#
# Keep notebook cleaner once finalised
import warnings
warnings.filterwarnings('ignore')
# Import packages
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Set up MatPlotLib
%matplotlib inline
# Change default colour scheme:
plt.style.use('seaborn-colorblind')
Collect the data#
We’ll use the data for large-vessel occlusions (LVOs) that are treated with mechanical thrombectomy (MT), using the same probability distributions that we’ve defined in this notebook.
Define the time of no effect \(t_{\mathrm{ne}}\) for LVOs treated with MT:
t_0 = 0.0 # hours
t_ne = 8.0 # hours
Load mRS distributions of probabilities in each bin, \(P(\mathrm{mRS}=x)\):
mrs_prob_all = pd.read_csv(
'./outcome_utilities/mrs_dist_probs_bins.csv', index_col='Stroke type')
mrs_prob_all
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| Stroke type | |||||||
| pre_stroke_nlvo | 0.582881 | 0.162538 | 0.103440 | 0.102223 | 0.041973 | 0.006945 | 0.000000 |
| pre_stroke_nlvo_ivt_deaths | 0.576469 | 0.160750 | 0.102302 | 0.101099 | 0.041511 | 0.006869 | 0.011000 |
| pre_stroke_lvo | 0.417894 | 0.142959 | 0.118430 | 0.164211 | 0.113775 | 0.042731 | 0.000000 |
| pre_stroke_lvo_ivt_deaths | 0.403644 | 0.138084 | 0.114392 | 0.158611 | 0.109895 | 0.041274 | 0.034100 |
| pre_stroke_lvo_mt_deaths | 0.401178 | 0.137241 | 0.113693 | 0.157643 | 0.109224 | 0.041022 | 0.040000 |
| no_treatment_nlvo | 0.197144 | 0.262856 | 0.120032 | 0.127736 | 0.147909 | 0.062025 | 0.082298 |
| no_effect_nlvo_ivt_deaths | 0.197271 | 0.262729 | 0.117583 | 0.124669 | 0.142991 | 0.059211 | 0.095546 |
| t0_treatment_nlvo_ivt | 0.429808 | 0.200192 | 0.108212 | 0.110215 | 0.080760 | 0.027113 | 0.043700 |
| no_treatment_lvo | 0.050000 | 0.079000 | 0.136000 | 0.164000 | 0.247000 | 0.135000 | 0.189000 |
| no_effect_lvo_ivt_deaths | 0.047898 | 0.075678 | 0.130282 | 0.157104 | 0.236614 | 0.129324 | 0.223100 |
| no_effect_lvo_mt_deaths | 0.047534 | 0.075104 | 0.129292 | 0.155911 | 0.234818 | 0.128342 | 0.229000 |
| t0_treatment_lvo_ivt | 0.112916 | 0.087084 | 0.127377 | 0.157380 | 0.213454 | 0.113231 | 0.188557 |
| t0_treatment_lvo_mt | 0.312767 | 0.121706 | 0.117593 | 0.157210 | 0.140622 | 0.062852 | 0.087250 |
Cumulative probabilities, \(P(\mathrm{mRS}\leq x))\):
mrs_cprob_all = pd.read_csv(
'./outcome_utilities/mrs_dist_probs_cumsum.csv', index_col='Stroke type')
mrs_cprob_all
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| Stroke type | |||||||
| pre_stroke_nlvo | 0.582881 | 0.745419 | 0.848859 | 0.951082 | 0.993055 | 1.000000 | 1.0 |
| pre_stroke_nlvo_ivt_deaths | 0.576469 | 0.737219 | 0.839522 | 0.940620 | 0.982131 | 0.989000 | 1.0 |
| pre_stroke_lvo | 0.417894 | 0.560853 | 0.679283 | 0.843494 | 0.957269 | 1.000000 | 1.0 |
| pre_stroke_lvo_ivt_deaths | 0.403644 | 0.541728 | 0.656119 | 0.814731 | 0.924626 | 0.965900 | 1.0 |
| pre_stroke_lvo_mt_deaths | 0.401178 | 0.538419 | 0.652112 | 0.809754 | 0.918978 | 0.960000 | 1.0 |
| no_treatment_nlvo | 0.197144 | 0.460000 | 0.580032 | 0.707768 | 0.855677 | 0.917702 | 1.0 |
| no_effect_nlvo_ivt_deaths | 0.197271 | 0.460000 | 0.577583 | 0.702252 | 0.845244 | 0.904454 | 1.0 |
| t0_treatment_nlvo_ivt | 0.429808 | 0.630000 | 0.738212 | 0.848427 | 0.929188 | 0.956300 | 1.0 |
| no_treatment_lvo | 0.050000 | 0.129000 | 0.265000 | 0.429000 | 0.676000 | 0.811000 | 1.0 |
| no_effect_lvo_ivt_deaths | 0.047898 | 0.123576 | 0.253858 | 0.410962 | 0.647576 | 0.776900 | 1.0 |
| no_effect_lvo_mt_deaths | 0.047534 | 0.122637 | 0.251930 | 0.407841 | 0.642658 | 0.771000 | 1.0 |
| t0_treatment_lvo_ivt | 0.112916 | 0.200000 | 0.327377 | 0.484757 | 0.698212 | 0.811443 | 1.0 |
| t0_treatment_lvo_mt | 0.312767 | 0.434474 | 0.552066 | 0.709276 | 0.849898 | 0.912750 | 1.0 |
Pick out the data we want here. In this example we will use the data for LVO patients treated with MT.
t0_st = 't0_treatment_lvo_mt'
ne_st = 'no_effect_lvo_mt_deaths'
mrs_prob_t0 = mrs_prob_all.loc[t0_st].values
mrs_prob_ne = mrs_prob_all.loc[ne_st].values
mrs_cprob_t0 = mrs_cprob_all.loc[t0_st].values
mrs_cprob_ne = mrs_cprob_all.loc[ne_st].values
Convert probability to odds:
mrs_odds_cumsum_t0 = mrs_cprob_t0 / (1.0 - mrs_cprob_t0)
mrs_odds_cumsum_ne = mrs_cprob_ne / (1.0 - mrs_cprob_ne)
# Manually set mRS<=6 values to infinity, since P(mRS<=6)=1:
mrs_odds_cumsum_t0[-1] = np.inf
mrs_odds_cumsum_ne[-1] = np.inf
Convert odds to log(odds):
mrs_logodds_cumsum_t0 = np.log(mrs_odds_cumsum_t0)
mrs_logodds_cumsum_ne = np.log(mrs_odds_cumsum_ne)
Plot the known data#
The following two cells define some colours and a plotting function that we’ll use here and later in the notebook:
colour_list = np.array([
'#0072B2', '#009E73', '#D55E00', '#CC79A7',
'#F0E442', '#56B4E9', 'DarkSlateGray'], dtype='<U13'
)
def plot_three_with_time(ylabels, data_lists):
fig, axs = plt.subplots(1,3, figsize=(15,5), gridspec_kw={'wspace':0.3})
for d,data_list in enumerate(data_lists):
ax = axs[d]
ax.grid()
d_t0 = data_list[0]
d_no = data_list[1]
for i in range(6):
ax.scatter([0.0,t_ne], [d_t0[i],d_no[i]],
color=colour_list[i], edgecolor='k',
label=f'$\leq${i}', zorder=3)
ax.set_ylabel(ylabels[d].split('\n')[0])
ax.set_title(ylabels[d], fontsize=15)
ax.set_xticks(np.arange(0,t_ne+1,1))
ax.set_xlabel('Onset to treatment time (hours)')
if d==1:
ax.legend(loc='upper center', bbox_to_anchor=[0.5, -0.15],
ncol=7, title='mRS')
axs[0].set_ylim(0.0, 1.0)
# axs[1].set_ylim(0.0, 11.5)
axs[2].axhline(0.0, color='k', linewidth=1)
return fig, axs
This cell produces the plot of probability, odds, and log(odds):
data_lists = [
[mrs_cprob_t0, mrs_cprob_ne],
[mrs_odds_cumsum_t0, mrs_odds_cumsum_ne],
[mrs_logodds_cumsum_t0, mrs_logodds_cumsum_ne]
]
ylabels = ['Probability', 'Odds', 'log(Odds)']
fig, axs = plot_three_with_time(ylabels, data_lists)
# plt.savefig('./images/time_varying_probs_odds_logodds_data-only.jpg',
# dpi=300, bbox_inches='tight')
plt.show()
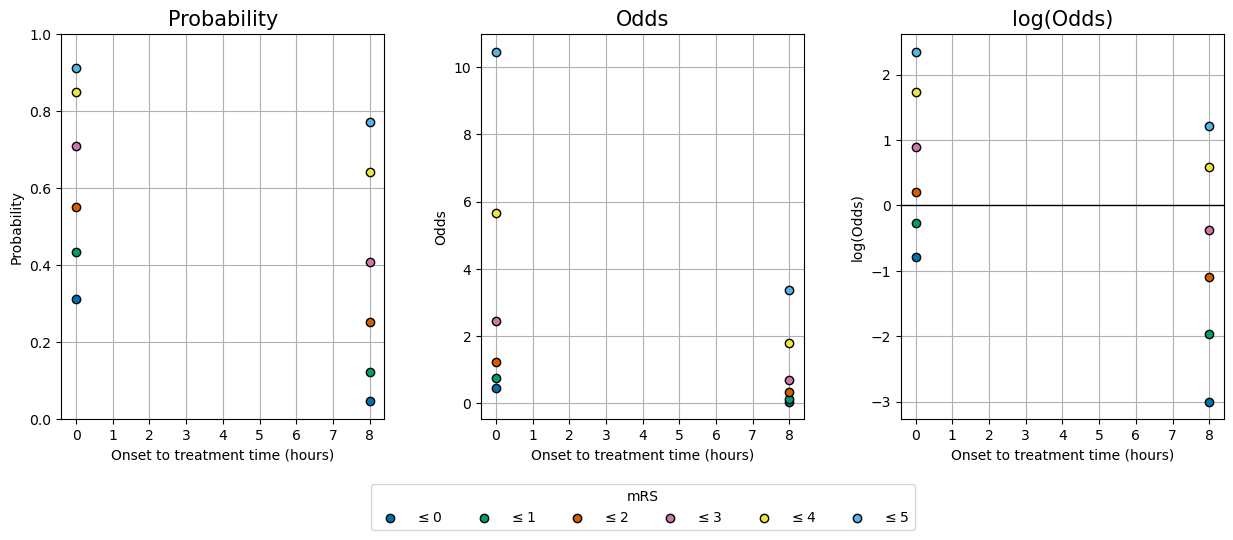
Now, for each graph we need to connect the two sets of data at \(t=0\) and the no-effect time by finding out how probability, odds and log(odds) vary with time.
Calculate log-odds proportions#
Pick out the data for mRS<=5 (the top points in the charts).
t0_logodds = mrs_logodds_cumsum_t0[5]
no_effect_logodds = mrs_logodds_cumsum_ne[5]
t0_logodds, no_effect_logodds
(2.347684459453209, 1.21396637000909)
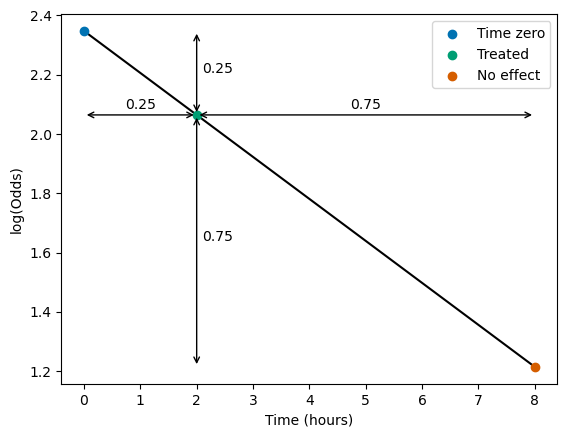
Pick a treatment time:
t_treat = 2.0 # hours
Find the proportion of time from onset to no effect:
frac_to_no_effect = (t_treat - t_0) / (t_ne - t_0)
frac_to_no_effect
0.25
Take this proportion from the starting to the end log-odds:
treated_logodds = t0_logodds - (frac_to_no_effect * (t0_logodds - no_effect_logodds))
treated_logodds
2.064254937092179
Plot this quickly to show how the proportions work:
plt.plot([t_0, t_ne], [t0_logodds, no_effect_logodds], color='k', zorder=0)
plt.scatter(t_0, t0_logodds, label='Time zero')
plt.scatter(t_treat, treated_logodds, label='Treated')
plt.scatter(t_ne, no_effect_logodds, label='No effect')
ad = dict(arrowstyle='<->')
# Horizontal arrows and labels:
plt.annotate('', xy=(t_0, treated_logodds),
xytext=(t_treat, treated_logodds),
arrowprops=ad)
plt.annotate(f'{frac_to_no_effect}',
xy=(0.5 * t_treat, treated_logodds * 1.01),
ha='center')
plt.annotate('', xy=(t_treat, treated_logodds),
xytext=(t_ne, treated_logodds),
arrowprops=ad)
plt.annotate(f'{1.0 - frac_to_no_effect}',
xy=(t_treat + 0.5 * (t_ne - t_treat), treated_logodds * 1.01),
ha='center')
# Vertical arrows and labels:
plt.annotate('', xy=(t_treat, t0_logodds),
xytext=(t_treat, treated_logodds), arrowprops=ad)
plt.annotate(f'{frac_to_no_effect}',
xy=(t_treat + 0.1, treated_logodds + 0.5 * (t0_logodds - treated_logodds)), ha='left')
plt.annotate('', xy=(t_treat, treated_logodds),
xytext=(t_treat, no_effect_logodds), arrowprops=ad)
plt.annotate(f'{1.0 - frac_to_no_effect}',
xy=(t_treat + 0.1, no_effect_logodds + 0.5 * (treated_logodds - no_effect_logodds)), ha='left')
plt.xlabel('Time (hours)')
plt.ylabel('log(Odds)')
plt.legend()
plt.show()
Finally, convert the treated log-odds to odds…
treated_odds = np.exp(treated_logodds)
treated_odds
6.838306928429508
… and these odds to probability:
treated_probs = treated_odds / (1 + treated_odds)
treated_probs
0.872421428615789
This is the probability of mRS\(\leq\)5 at the chosen treatment time.
By repeating this for all mRS values, we can find the mRS distribution at this treatment time:
treated_logodds = (
mrs_logodds_cumsum_t0 - (frac_to_no_effect * (mrs_logodds_cumsum_t0 - mrs_logodds_cumsum_ne))
)
treated_odds = np.exp(treated_logodds)
treated_probs = treated_odds / (1 + treated_odds)
treated_probs
array([0.16574163, 0.28850692, 0.431069 , 0.60290449, 0.78646085,
0.87242143, nan])
The final value is nan, Not A Number, because mRS\(\leq\)6 always has a probability of 1 and so cannot be converted into log-odds and back. When working with mRS\(\leq\)6 we can just manually change any invalid probabilities to 1 at the end of the calculations.
stroke-outcome package#
The stroke-outcome package includes the following function to calculate the probability at a given treatment time.
from stroke_outcome.outcome_utilities import calculate_mrs_dist_at_treatment_time
Display the contents of the function:
calculate_mrs_dist_at_treatment_time??
Signature:
calculate_mrs_dist_at_treatment_time(
time_to_treatment_mins: Union[numpy.__array_like._SupportsArray[numpy.dtype[Any]], numpy.__nested_sequence._NestedSequence[numpy.__array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy.__nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]],
time_no_effect_mins: float,
t0_logodds: Union[numpy.__array_like._SupportsArray[numpy.dtype[Any]], numpy.__nested_sequence._NestedSequence[numpy.__array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy.__nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]],
no_effect_logodds: Union[numpy.__array_like._SupportsArray[numpy.dtype[Any]], numpy.__nested_sequence._NestedSequence[numpy.__array_like._SupportsArray[numpy.dtype[Any]]], bool, int, float, complex, str, bytes, numpy.__nested_sequence._NestedSequence[Union[bool, int, float, complex, str, bytes]]],
final_value_is_mrs6: bool = True,
)
Source:
def calculate_mrs_dist_at_treatment_time(
time_to_treatment_mins: npt.ArrayLike,
time_no_effect_mins: float,
t0_logodds: npt.ArrayLike,
no_effect_logodds: npt.ArrayLike,
final_value_is_mrs6: bool = True
):
"""
Calculate the mRS distribution at arbitrary treatment time(s).
If the input log-odds arrays contain one value per mRS score,
then the output treated probabilities will make the
mRS distribution at the treatment time.
The new distributions are created by calculating log-odds at
the treatment time. For each mRS band, the method is:
l | Draw a straight line between the log-odds
o |x1 treated at time zero and the time of no effect.
g | \ at "o" Then the log-odds at the chosen treatment
o | \ time lies on this line.
d | o
d | \
s |__________x2__
time
The (x,y) coordinates of the two points are:
x1: (0, t0_logodds)
x2: (time_no_effect_mins, no_effect_logodds)
o: (time_to_treatment_mins, treated_logodds)
The log-odds are then translated to odds and probability:
odds = exp(log-odds)
prob = odds / (1 + odds)
Inputs:
-------
time_to_treatment_mins - np.array. The time to treatment in
minutes. Can contain multiple values so
long as the array shape is (X, 1).
time_no_effect_mins - float. Time of no effect in minutes.
t0_logodds - np.array. Log-odds at time zero. Can
provide one value per mRS score.
no_effect_logodds - np.array. Log-odds at time of no
effect. Can provide one value per mRS
score.
final_value_is_mrs6 - bool. Whether the final logodds value
is for mRS<=6. If True, the final
probabilities are all set to 1 to replace
the default Not A Number values.
Returns:
--------
treated_probs - np.array. mRS probability distribution at the
treatment time(s).
treated_odds - np.array. As above, but converted to odds.
treated_logodds - np.array. As above, but converted to log-odds.
"""
# Calculate fraction of time to no effect passed
frac_to_no_effect = time_to_treatment_mins / time_no_effect_mins
# Combine t=0 and no effect distributions based on time passed
treated_logodds = ((frac_to_no_effect * no_effect_logodds) +
((1 - frac_to_no_effect) * t0_logodds))
# Convert to odds and probabilties
treated_odds = np.exp(treated_logodds)
treated_probs = treated_odds / (1 + treated_odds)
if final_value_is_mrs6 is True:
# Manually set all of the probabilities for mRS<=6 to be 1
# as the logodds calculation returns NaN.
if len(treated_probs.shape) == 1:
treated_probs[-1] = 1.0
else:
treated_probs[:, -1] = 1.0
return treated_probs, treated_odds, treated_logodds
File: ~/miniconda3/lib/python3.9/site-packages/stroke_outcome/outcome_utilities.py
Type: function
.
Check that this gives the same results as for earlier in this document:
treated_probs_before = treated_probs.copy()
treated_probs, treated_odds, treated_logodds = calculate_mrs_dist_at_treatment_time(
time_to_treatment_mins = t_treat,
time_no_effect_mins = t_ne,
t0_logodds = mrs_logodds_cumsum_t0,
no_effect_logodds = mrs_logodds_cumsum_ne,
final_value_is_mrs6 = True
)
treated_probs
array([0.16574163, 0.28850692, 0.431069 , 0.60290449, 0.78646085,
0.87242143, 1. ])
treated_probs == treated_probs_before
array([ True, True, True, True, True, True, True])
Plot the time distributions#
The following cell draws the same graphs as before, and now adds the variations with time.
The function from the stroke-outcome package is used to calculate the probability distributions at very many times between time zero and the time of no effect so that on the final graphs it looks like a continuous change.
ylabels = ['Probability', 'Odds', 'log(Odds)']
# Use these logodds:
t0_logodds = mrs_logodds_cumsum_t0
no_effect_logodds = mrs_logodds_cumsum_ne
# This time-of-no-effect:
t_ne = 8.0 # hours
# Plot over this time range:
t = np.arange(0, t_ne*60.0 + 0.1, 0.1)
# Reshape the arrays to allow for multiple treatment times.
time_to_treatment_mins = t.reshape(len(t), 1)
no_effect_logodds = no_effect_logodds.reshape(1, len(no_effect_logodds))
t0_logodds = t0_logodds.reshape(1, len(t0_logodds))
treated_probs, treated_odds, treated_logodds = calculate_mrs_dist_at_treatment_time(
time_to_treatment_mins = time_to_treatment_mins,
time_no_effect_mins = t_ne * 60.0,
t0_logodds = t0_logodds,
no_effect_logodds = no_effect_logodds,
final_value_is_mrs6 = True
)
# Make the same plot as earlier...
fig, axs = plot_three_with_time(ylabels, data_lists)
# ... and now add lines to connect the starting data points:
for d, ax in enumerate(axs):
for i in range(6):
if d==0: # Probability:
yvals = treated_probs[:, i]
elif d==1: # Odds:
yvals = treated_odds[:, i]
else: # log(odds):
yvals = treated_logodds[:, i]
ax.plot(t / 60.0, yvals, color=colour_list[i], zorder=2)
# plt.savefig('./images/time_varying_probs_odds_logodds.jpg',
# dpi=300, bbox_inches='tight')
plt.show()
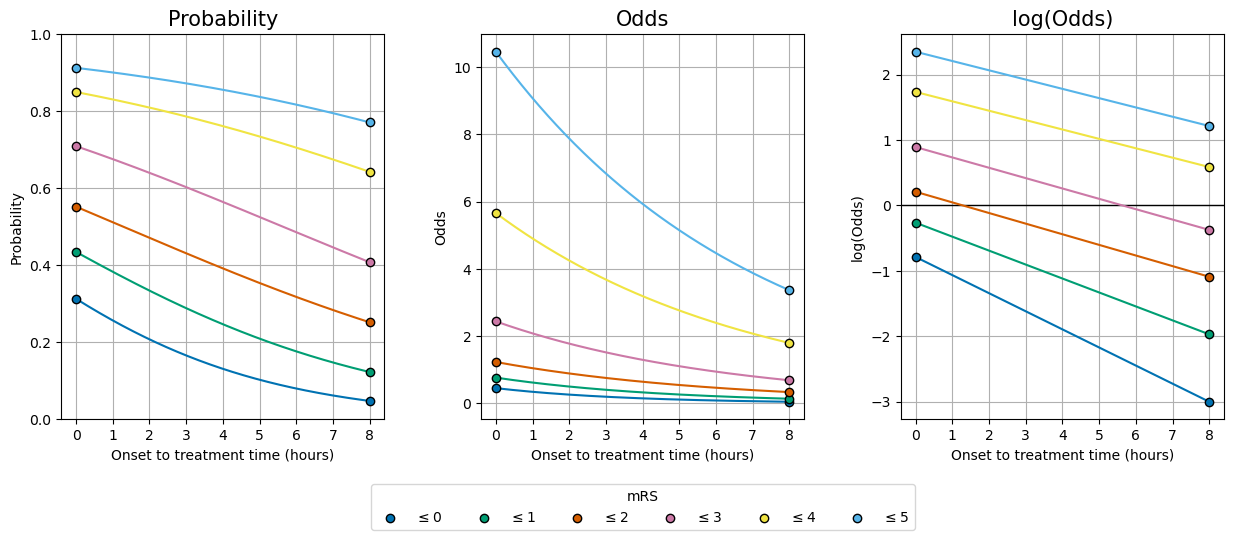
Success! We started with the data at t=0 and t=\(t_{\mathrm{ne}}\) (8 hours) and have calculated the lines in between.
References#
Emberson J, Lees KR, Lyden P, et al. Effect of treatment delay, age, and stroke severity on the effects of intravenous thrombolysis with alteplase for acute ischaemic stroke: A meta-analysis of individual patient data from randomised trials. The Lancet 2014;384:1929–35. doi:10.1016/S0140-6736(14)60584-5
Fransen PSS, Berkhemer OA, Lingsma HF, et al. Time to Reperfusion and Treatment Effect for Acute Ischemic Stroke: A Randomized Clinical Trial. JAMA Neurol 2016;73:190–6. doi:10.1001/jamaneurol.2015.3886