Mapping outcomes#
This notebook creates maps of England and Wales to show the variation in outcomes from the different scenarios calculated earlier.
Plain English summary#
This notebook takes the outcome data we saved earlier and creates maps of England and Wales.
Aim#
Create maps to show:
change in added utility compared with no treatment
change in mean mRS score compared with no treatment
change in proportion of the population with an mRS score of 2 or less
for each of these cohorts:
nLVO treated with IVT
LVO treated with mixed methods
the treated ischaemic population
for each of these scenarios:
drip-and-ship
mothership
Method#
We load LSOA shapes from the Office for National Statistics and merge this geography data into the outcome data. Then we draw the shape of each LSOA with a colour that depends on the outcome value. All of the LSOAs on a map use the same range of colours so that they can be directly compared.
Import packages#
# import contextily as ctx
import geopandas
import numpy as np
import pandas as pd
import os
# For plotting:
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import stroke_maps.load_data
import stroke_maps.catchment
import stroke_maps.geo # to make catchment area geometry
pd.set_option('display.max_rows', 150)
dir_output = 'output'
limit_to_england = False
Load data#
Load shape file#
lsoa_gdf = stroke_maps.load_data.lsoa_geography()
lsoa_gdf = lsoa_gdf.to_crs('EPSG:27700')
lsoa_gdf.head(3)
| OBJECTID | LSOA11CD | LSOA11NM | LSOA11NMW | BNG_E | BNG_N | LONG | LAT | Shape__Area | Shape__Length | GlobalID | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | E01000001 | City of London 001A | City of London 001A | 532129 | 181625 | -0.09706 | 51.51810 | 157794.481079 | 1685.391778 | b12173a3-5423-4672-a5eb-f152d2345f96 | POLYGON ((532282.642 181906.500, 532248.262 18... |
| 1 | 2 | E01000002 | City of London 001B | City of London 001B | 532480 | 181699 | -0.09197 | 51.51868 | 164882.427628 | 1804.828196 | 90274dc4-f785-4afb-95cd-7cc1fc9a2cad | POLYGON ((532746.826 181786.896, 532248.262 18... |
| 2 | 3 | E01000003 | City of London 001C | City of London 001C | 532245 | 182036 | -0.09523 | 51.52176 | 42219.805717 | 909.223277 | 7e89d0ba-f186-45fb-961c-8f5ffcd03808 | POLYGON ((532293.080 182068.426, 532419.605 18... |
# Load country outline
if limit_to_england:
outline = stroke_maps.load_data.england_outline()
else:
outline = stroke_maps.load_data.englandwales_outline()
outline
| country | OBJECTID | ctry11cd | ctry11cdo | ctry11nm | ctry11nmw | GlobalID | geometry | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | E92000001 | 921 | England | Lloegr | 27bbf7ca-99bd-4fe8-87a1-d498d48e3084 | MULTIPOLYGON (((83994.599 5397.099, 84001.300 ... |
Load hospital info#
Load in the stroke unit coordinates and merge in the services information:
df_units = stroke_maps.load_data.stroke_unit_region_lookup()
df_units.head(3).T
| postcode | SY231ER | CB20QQ | L97AL |
|---|---|---|---|
| stroke_team | Bronglais Hospital (Aberystwyth) | Addenbrooke's Hospital, Cambridge | University Hospital Aintree, Liverpool |
| short_code | AB | AD | AI |
| ssnap_name | Bronglais Hospital | Addenbrooke's Hospital | University Hospital Aintree |
| use_ivt | 1 | 1 | 1 |
| use_mt | 0 | 1 | 1 |
| use_msu | 0 | 1 | 1 |
| transfer_unit_postcode | nearest | nearest | nearest |
| lsoa | Ceredigion 002A | Cambridge 013D | Liverpool 005A |
| lsoa_code | W01000512 | E01017995 | E01006654 |
| region | Hywel Dda University Health Board | NHS Cambridgeshire and Peterborough ICB - 06H | NHS Cheshire and Merseyside ICB - 99A |
| region_code | W11000025 | E38000260 | E38000101 |
| region_type | LHB | SICBL | SICBL |
| country | Wales | England | England |
| icb | NaN | NHS Cambridgeshire and Peterborough Integrated... | NHS Cheshire and Merseyside Integrated Care Board |
| icb_code | NaN | E54000056 | E54000008 |
| isdn | NaN | East of England (South) | Cheshire and Merseyside |
hospitals_gdf = stroke_maps.load_data.stroke_unit_coordinates()
hospitals_gdf = pd.merge(
hospitals_gdf, df_units[['use_ivt', 'use_mt']],
left_index=True, right_index=True, how='right'
)
hospitals_gdf.head(3)
| BNG_E | BNG_N | Latitude | Longitude | geometry | use_ivt | use_mt | |
|---|---|---|---|---|---|---|---|
| postcode | |||||||
| SY231ER | 259208 | 281805 | 52.416068 | -4.071578 | POINT (259208.000 281805.000) | 1 | 0 |
| CB20QQ | 546375 | 254988 | 52.173741 | 0.139114 | POINT (546375.000 254988.000) | 1 | 1 |
| L97AL | 338020 | 397205 | 53.467918 | -2.935131 | POINT (338020.000 397205.000) | 1 | 1 |
Load LSOA model output data#
lsoa_data = pd.read_csv(os.path.join(dir_output, 'cohort_outcomes_weighted.csv'))
lsoa_data.head(3).T
| 0 | 1 | 2 | |
|---|---|---|---|
| lsoa | Adur 001A | Adur 001B | Adur 001C |
| closest_ivt_time | 17.6 | 18.7 | 17.6 |
| closest_ivt_unit | BN25BE | BN25BE | BN112DH |
| closest_mt_time | 17.6 | 18.7 | 19.8 |
| closest_mt_unit | BN25BE | BN25BE | BN25BE |
| transfer_mt_time | 0.0 | 0.0 | 31.6 |
| transfer_mt_unit | BN25BE | BN25BE | BN25BE |
| mt_transfer_required | False | False | True |
| ivt_drip_ship | 107.6 | 108.7 | 107.6 |
| mt_drip_ship | 167.6 | 168.7 | 259.2 |
| ivt_mothership | 107.6 | 108.7 | 109.8 |
| mt_mothership | 167.6 | 168.7 | 169.8 |
| drip_ship_nlvo_ivt_added_utility | 0.11685 | 0.11641 | 0.11685 |
| drip_ship_nlvo_ivt_mean_mrs | 1.63653 | 1.63913 | 1.63653 |
| drip_ship_nlvo_ivt_mrs_less_equal_2 | 0.70649 | 0.706 | 0.70649 |
| drip_ship_nlvo_ivt_mrs_shift | -0.64347 | -0.64087 | -0.64347 |
| drip_ship_nlvo_ivt_added_mrs_less_equal_2 | 0.12649 | 0.126 | 0.12649 |
| drip_ship_lvo_ivt_added_utility | 0.05872 | 0.05841 | 0.05872 |
| drip_ship_lvo_ivt_mean_mrs | 3.34655 | 3.34823 | 3.34655 |
| drip_ship_lvo_ivt_mrs_less_equal_2 | 0.32879 | 0.32847 | 0.32879 |
| drip_ship_lvo_ivt_mrs_shift | -0.29345 | -0.29177 | -0.29345 |
| drip_ship_lvo_ivt_added_mrs_less_equal_2 | 0.06379 | 0.06347 | 0.06379 |
| drip_ship_lvo_ivt_mt_added_utility | 0.16361 | 0.16295 | 0.10928 |
| drip_ship_lvo_ivt_mt_mean_mrs | 2.81136 | 2.81494 | 3.10174 |
| drip_ship_lvo_ivt_mt_mrs_less_equal_2 | 0.43807 | 0.43736 | 0.37981 |
| drip_ship_lvo_ivt_mt_mrs_shift | -0.82864 | -0.82506 | -0.53826 |
| drip_ship_lvo_ivt_mt_added_mrs_less_equal_2 | 0.17307 | 0.17236 | 0.11481 |
| drip_ship_lvo_mt_added_utility | 0.16361 | 0.16295 | 0.10928 |
| drip_ship_lvo_mt_mean_mrs | 2.81136 | 2.81494 | 3.10174 |
| drip_ship_lvo_mt_mrs_less_equal_2 | 0.43807 | 0.43736 | 0.37981 |
| drip_ship_lvo_mt_mrs_shift | -0.82864 | -0.82506 | -0.53826 |
| drip_ship_lvo_mt_added_mrs_less_equal_2 | 0.17307 | 0.17236 | 0.11481 |
| mothership_nlvo_ivt_added_utility | 0.11685 | 0.11641 | 0.11596 |
| mothership_nlvo_ivt_mean_mrs | 1.63653 | 1.63913 | 1.64173 |
| mothership_nlvo_ivt_mrs_less_equal_2 | 0.70649 | 0.706 | 0.70551 |
| mothership_nlvo_ivt_mrs_shift | -0.64347 | -0.64087 | -0.63827 |
| mothership_nlvo_ivt_added_mrs_less_equal_2 | 0.12649 | 0.126 | 0.12551 |
| mothership_lvo_ivt_added_utility | 0.05872 | 0.05841 | 0.05811 |
| mothership_lvo_ivt_mean_mrs | 3.34655 | 3.34823 | 3.3499 |
| mothership_lvo_ivt_mrs_less_equal_2 | 0.32879 | 0.32847 | 0.32815 |
| mothership_lvo_ivt_mrs_shift | -0.29345 | -0.29177 | -0.2901 |
| mothership_lvo_ivt_added_mrs_less_equal_2 | 0.06379 | 0.06347 | 0.06315 |
| mothership_lvo_ivt_mt_added_utility | 0.16361 | 0.16295 | 0.16229 |
| mothership_lvo_ivt_mt_mean_mrs | 2.81136 | 2.81494 | 2.81852 |
| mothership_lvo_ivt_mt_mrs_less_equal_2 | 0.43807 | 0.43736 | 0.43664 |
| mothership_lvo_ivt_mt_mrs_shift | -0.82864 | -0.82506 | -0.82148 |
| mothership_lvo_ivt_mt_added_mrs_less_equal_2 | 0.17307 | 0.17236 | 0.17164 |
| mothership_lvo_mt_added_utility | 0.16361 | 0.16295 | 0.16229 |
| mothership_lvo_mt_mean_mrs | 2.81136 | 2.81494 | 2.81852 |
| mothership_lvo_mt_mrs_less_equal_2 | 0.43807 | 0.43736 | 0.43664 |
| mothership_lvo_mt_mrs_shift | -0.82864 | -0.82506 | -0.82148 |
| mothership_lvo_mt_added_mrs_less_equal_2 | 0.17307 | 0.17236 | 0.17164 |
| drip_ship_lvo_mix_added_utility | 0.15548 | 0.15485 | 0.10536 |
| drip_ship_lvo_mix_mrs_less_equal_2 | 0.4296 | 0.42892 | 0.37586 |
| drip_ship_lvo_mix_mrs_shift | -0.78717 | -0.78373 | -0.51929 |
| drip_ship_lvo_mix_added_mrs_less_equal_2 | 0.1646 | 0.16392 | 0.11086 |
| mothership_lvo_mix_added_utility | 0.15548 | 0.15485 | 0.15422 |
| mothership_lvo_mix_mrs_less_equal_2 | 0.4296 | 0.42892 | 0.42823 |
| mothership_lvo_mix_mrs_shift | -0.78717 | -0.78373 | -0.7803 |
| mothership_lvo_mix_added_mrs_less_equal_2 | 0.1646 | 0.16392 | 0.16323 |
| drip_ship_weighted_added_utility | 0.03392 | 0.03379 | 0.02764 |
| drip_ship_weighted_mrs_less_equal_2 | 0.14107 | 0.14092 | 0.13432 |
| drip_ship_weighted_mrs_shift | -0.17815 | -0.1774 | -0.14454 |
| drip_ship_weighted_added_mrs_less_equal_2 | 0.03626 | 0.03611 | 0.02951 |
| mothership_weighted_added_utility | 0.03392 | 0.03379 | 0.03366 |
| mothership_weighted_mrs_less_equal_2 | 0.14107 | 0.14092 | 0.14077 |
| mothership_weighted_mrs_shift | -0.17815 | -0.1774 | -0.17665 |
| mothership_weighted_added_mrs_less_equal_2 | 0.03626 | 0.03611 | 0.03597 |
| drip_ship_weighted_treated_added_utility | 0.13633 | 0.13579 | 0.11106 |
| drip_ship_weighted_treated_mrs_less_equal_2 | 0.56689 | 0.5663 | 0.53979 |
| drip_ship_weighted_treated_mrs_shift | -0.71592 | -0.7129 | -0.58086 |
| drip_ship_weighted_treated_added_mrs_less_equal_2 | 0.14571 | 0.14512 | 0.11861 |
| mothership_weighted_treated_added_utility | 0.13633 | 0.13579 | 0.13525 |
| mothership_weighted_treated_mrs_less_equal_2 | 0.56689 | 0.5663 | 0.56571 |
| mothership_weighted_treated_mrs_shift | -0.71592 | -0.7129 | -0.70988 |
| mothership_weighted_treated_added_mrs_less_equal_2 | 0.14571 | 0.14512 | 0.14453 |
# Merge with shape file
lsoa_data_gdf = lsoa_gdf.merge(lsoa_data, left_on='LSOA11NM', right_on='lsoa', how='right')
lsoa_data_gdf.head()
| OBJECTID | LSOA11CD | LSOA11NM | LSOA11NMW | BNG_E | BNG_N | LONG | LAT | Shape__Area | Shape__Length | ... | mothership_weighted_mrs_shift | mothership_weighted_added_mrs_less_equal_2 | drip_ship_weighted_treated_added_utility | drip_ship_weighted_treated_mrs_less_equal_2 | drip_ship_weighted_treated_mrs_shift | drip_ship_weighted_treated_added_mrs_less_equal_2 | mothership_weighted_treated_added_utility | mothership_weighted_treated_mrs_less_equal_2 | mothership_weighted_treated_mrs_shift | mothership_weighted_treated_added_mrs_less_equal_2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 30557.0 | E01031349 | Adur 001A | Adur 001A | 524915.0 | 105607.0 | -0.22737 | 50.83651 | 3.641032e+05 | 3054.751704 | ... | -0.17815 | 0.03626 | 0.13633 | 0.56689 | -0.71592 | 0.14571 | 0.13633 | 0.56689 | -0.71592 | 0.14571 |
| 1 | 30558.0 | E01031350 | Adur 001B | Adur 001B | 524825.0 | 106265.0 | -0.22842 | 50.84244 | 2.921732e+05 | 2977.102897 | ... | -0.17740 | 0.03611 | 0.13579 | 0.56630 | -0.71290 | 0.14512 | 0.13579 | 0.56630 | -0.71290 | 0.14512 |
| 2 | 30559.0 | E01031351 | Adur 001C | Adur 001C | 523053.0 | 108004.0 | -0.25300 | 50.85845 | 5.281768e+06 | 11671.349143 | ... | -0.17665 | 0.03597 | 0.11106 | 0.53979 | -0.58086 | 0.11861 | 0.13525 | 0.56571 | -0.70988 | 0.14453 |
| 3 | 30560.0 | E01031352 | Adur 001D | Adur 001D | 524141.0 | 106299.0 | -0.23812 | 50.84290 | 2.452292e+05 | 2134.908586 | ... | -0.17665 | 0.03597 | 0.11106 | 0.53979 | -0.58086 | 0.11861 | 0.13525 | 0.56571 | -0.70988 | 0.14453 |
| 4 | 30578.0 | E01031370 | Adur 001E | Adur 001E | 523561.0 | 105916.0 | -0.24649 | 50.83958 | 2.402445e+05 | 2447.096939 | ... | -0.17665 | 0.03597 | 0.11159 | 0.54036 | -0.58379 | 0.11918 | 0.13525 | 0.56571 | -0.70988 | 0.14453 |
5 rows × 88 columns
Calculate difference between Mothership and Drip and Ship#
cohort_names = ['nlvo_ivt', 'lvo_mix', 'weighted_treated']
outcome_names = ['added_utility', 'added_mrs_less_equal_2', 'mrs_shift']
cols_diff = [f'{c}_{o}_mothership_minus_dripship' for c in cohort_names for o in outcome_names]
cols_moth = [f'mothership_{c}_{o}' for c in cohort_names for o in outcome_names]
cols_drip = [f'drip_ship_{c}_{o}' for c in cohort_names for o in outcome_names]
lsoa_data_gdf[cols_diff] = lsoa_data_gdf[cols_moth].values - lsoa_data_gdf[cols_drip].values
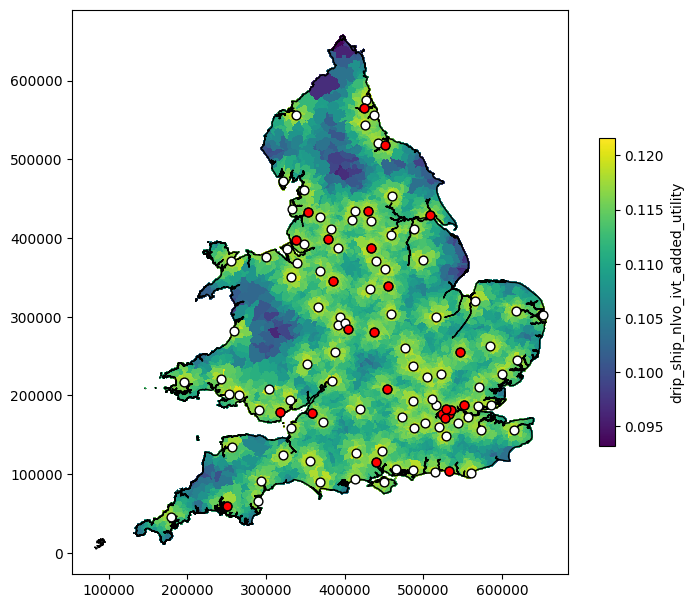
Basic plot#
col = 'drip_ship_nlvo_ivt_added_utility'
# Figure setup:
fig, ax = plt.subplots(figsize=(8, 8))
# Plot data
lsoa_data_gdf.plot(
ax=ax, # Set which axes to use for plot
column=col, # Column to apply colour
antialiased=False, # Avoids artefact boundry lines
edgecolor='face', # Make LSOA boundary same colour as area
# Adjust size of colourmap key, and add label
legend_kwds={'shrink':0.5, 'label':col},
legend=True, # Set to display legend
)
# Add country border
outline.plot(ax=ax, edgecolor='k', facecolor='None', linewidth=1.0)
# Add hospitals
mask = hospitals_gdf['use_ivt'] == 1
hospitals_gdf[mask].plot(ax=ax, edgecolor='k', facecolor='w', markersize=40, marker='o')
mask = hospitals_gdf['use_mt'] == 1
hospitals_gdf[mask].plot(ax=ax, edgecolor='k', facecolor='r', markersize=40, marker='o')
plt.show()
Plots#
Define functions#
These functions find shared colour limits across multiple columns of data:
def find_vlims_scenarios(lsoa_data_gdf, data_field, cohorts):
scenarios = ['drip_ship', 'mothership']
# Colour limits for separate scenarios:
cols = [f'{s}_{c}_{data_field}' for s in scenarios for c in cohorts]
# Find maximum of data
vmin = np.min([lsoa_data_gdf[col].min() for col in cols])
vmax = np.max([lsoa_data_gdf[col].max() for col in cols])
return vmin, vmax
def find_vlims_diff(lsoa_data_gdf, data_field, cohorts):
# Colour limits for difference maps:
cols = [f'{c}_{data_field}_mothership_minus_dripship' for c in cohorts]
# Find absolute maximum of data extent
vmax = np.max((np.abs([lsoa_data_gdf[col].min() for col in cols]),
np.abs([lsoa_data_gdf[col].max() for col in cols])))
vmin = -vmax
return vmin, vmax
This function plots the maps:
def plot_data(axs, cax, cax_diff, axs_cols, axs_params, colour_params):
for row in range(len(axs_cols)):
for col in range(len(axs_cols[row])):
# Axis to plot on:
ax = axs[row, col]
# Data information:
col_data = axs_cols[row][col]
# Colour information:
colour_params_type = axs_params[row][col]
params = colour_params[colour_params_type]
cbar_ax = cax if colour_params_type == 'shared' else cax_diff
# Plot data
lsoa_data_gdf.plot(
ax=ax,
column=col_data, # Column to apply colour
antialiased=False, # Avoids artefact boundry lines
edgecolor='face', # Make LSOA boundry same colour as area
vmin=params['vmin'], # Manual scale min (remove to make automatic)
vmax=params['vmax'], # Manual scale max (remove to make automatic)
cmap=params['cmap'], # Colour map to use
# Adjust size of colourmap key, and add label
legend_kwds={'shrink':0.5, 'label':params['cbar_label']},
legend=True, # Set to display legend
cax=cbar_ax
)
These functions set up the figure and standard formatting across all axes:
def set_up_fig_nine():
fig = plt.figure(figsize=(14, 18))
# Set up GridSpec so that each map subplot takes up two gs subplots
# in height. This lets the shared colourbar sit midway up two
# subplots instead of being offset or twice the height of the other.
gs = GridSpec(7, 4, width_ratios=[1, 1, 1, 0.05], wspace=0.0, hspace=0.0)
axs = np.array([
[plt.subplot(gs[0:2, 0]), plt.subplot(gs[0:2, 1]), plt.subplot(gs[0:2, 2])],
[plt.subplot(gs[2:4, 0]), plt.subplot(gs[2:4, 1]), plt.subplot(gs[2:4, 2])],
[plt.subplot(gs[4:6, 0]), plt.subplot(gs[4:6, 1]), plt.subplot(gs[4:6, 2])],
])
cax = plt.subplot(gs[1:3, -1])
cax_diff = plt.subplot(gs[4:6, -1])
return fig, axs, cax, cax_diff
def set_up_axis_and_extras(ax, outline, hospitals_gdf):
# Add country border
outline.plot(ax=ax, edgecolor='k', facecolor='None', linewidth=1.0)
# Add hospitals
mask = hospitals_gdf['use_ivt'] == 1
hospitals_gdf[mask].plot(ax=ax, edgecolor='k', facecolor='w', markersize=40, marker='o')
mask = hospitals_gdf['use_mt'] == 1
hospitals_gdf[mask].plot(ax=ax, edgecolor='k', facecolor='r', markersize=40, marker='o')
# ax.set_axis_off() # Turn of axis line and numbers
ax.set_xticks([])
ax.set_yticks([])
for spine in ['top', 'bottom', 'left', 'right']:
ax.spines[spine].set_visible(False)
# Tighten map around mainland England and Wales
ax.set_xlim(120000)
return ax
This function makes the plots for nine maps together:
def plot_nine(
lsoa_data_gdf,
axs_cols,
axs_params,
colour_params,
hospitals_gdf,
outline,
col_titles=[],
row_titles=[],
savename=''
):
fig, axs, cax, cax_diff = set_up_fig_nine()
plot_data(axs, cax, cax_diff, axs_cols, axs_params, colour_params)
for ax in axs.flatten():
ax = set_up_axis_and_extras(ax, outline, hospitals_gdf)
for i, row_title in enumerate(row_titles):
axs[i, 0].set_ylabel(row_title, rotation=0, fontsize=14, labelpad=50.0)#, ha='right')
for i, col_title in enumerate(col_titles):
axs[0, i].set_xlabel(col_title, fontsize=14)
axs[0, i].xaxis.set_label_position('top')
plt.tight_layout(pad=1)
if len(savename) > 0:
plt.savefig(savename, dpi=300, bbox_inches='tight')
plt.show()
Draw the plots#
Settings for the nine-in-one plot:
col_titles = ['nLVO', 'LVO', 'Treated ischaemic population']
row_titles = ['Drip and ship', 'Mothership', 'Advantage of\n mothership']
cohorts = ['nlvo_ivt', 'lvo_mix', 'weighted_treated']
axs_params = [
['shared'] * 3,
['shared'] * 3,
['diff'] * 3,
]
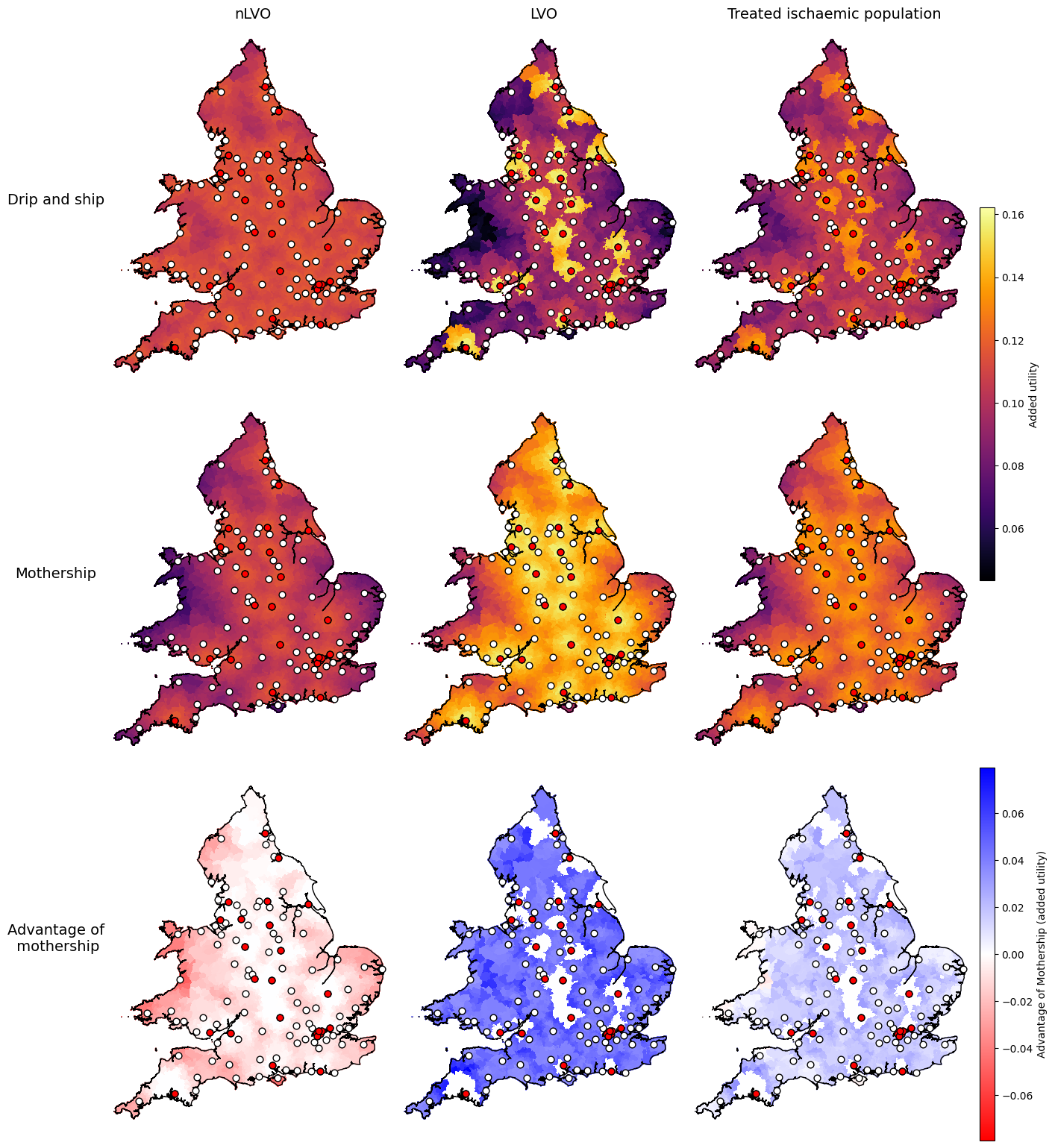
Settings for added utility:
data_field = 'added_utility'
vmin_scenarios, vmax_scenarios = find_vlims_scenarios(lsoa_data_gdf, data_field, cohorts)
vmin_diff, vmax_diff = find_vlims_diff(lsoa_data_gdf, data_field, cohorts)
params_added_utility = {
'shared': {
'vmin': vmin_scenarios,
'vmax': vmax_scenarios,
'cmap': 'inferno',
'cbar_label': 'Added utility'
},
'diff': {
'vmin': vmin_diff,
'vmax': vmax_diff,
'cmap': 'bwr_r',
'cbar_label': 'Advantage of Mothership (added utility)'
},
}
axs_cols_added_utility = [
# First row:
[f'drip_ship_nlvo_ivt_{data_field}',
f'drip_ship_lvo_mix_{data_field}',
f'drip_ship_weighted_treated_{data_field}'],
# Second row:
[f'mothership_nlvo_ivt_{data_field}',
f'mothership_lvo_mix_{data_field}',
f'mothership_weighted_treated_{data_field}'],
# Third row:
[f'nlvo_ivt_{data_field}_mothership_minus_dripship',
f'lvo_mix_{data_field}_mothership_minus_dripship',
f'weighted_treated_{data_field}_mothership_minus_dripship'],
]
plot_nine(
lsoa_data_gdf,
axs_cols_added_utility,
axs_params,
params_added_utility,
hospitals_gdf,
outline,
col_titles,
row_titles,
savename=os.path.join(dir_output, f'{data_field}_nine_in_one.jpg')
)
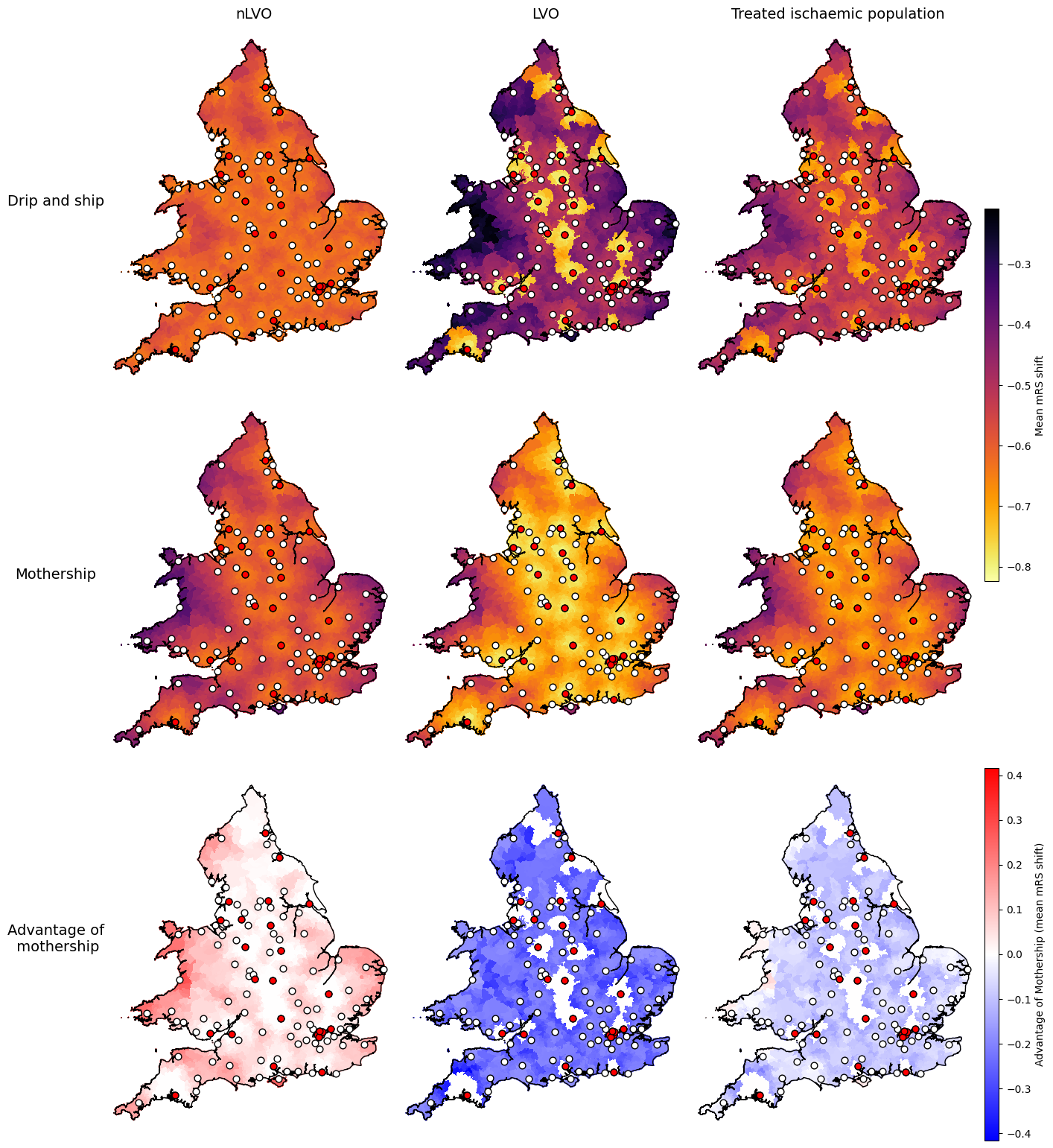
Settings for mRS shift:
data_field = 'mrs_shift'
vmin_scenarios, vmax_scenarios = find_vlims_scenarios(lsoa_data_gdf, data_field, cohorts)
vmin_diff, vmax_diff = find_vlims_diff(lsoa_data_gdf, data_field, cohorts)
params_mrs_shift = {
'shared': {
'vmin': vmin_scenarios,
'vmax': vmax_scenarios,
'cmap': 'inferno_r',
'cbar_label': 'Mean mRS shift'
},
'diff': {
'vmin': vmin_diff,
'vmax': vmax_diff,
'cmap': 'bwr',
'cbar_label': 'Advantage of Mothership (mean mRS shift)'
},
}
axs_cols_mrs_shift = [
# First row:
[f'drip_ship_nlvo_ivt_{data_field}',
f'drip_ship_lvo_mix_{data_field}',
f'drip_ship_weighted_treated_{data_field}'],
# Second row:
[f'mothership_nlvo_ivt_{data_field}',
f'mothership_lvo_mix_{data_field}',
f'mothership_weighted_treated_{data_field}'],
# Third row:
[f'nlvo_ivt_{data_field}_mothership_minus_dripship',
f'lvo_mix_{data_field}_mothership_minus_dripship',
f'weighted_treated_{data_field}_mothership_minus_dripship'],
]
plot_nine(
lsoa_data_gdf,
axs_cols_mrs_shift,
axs_params,
params_mrs_shift,
hospitals_gdf,
outline,
col_titles,
row_titles,
savename=os.path.join(dir_output, f'{data_field}_nine_in_one.jpg')
)
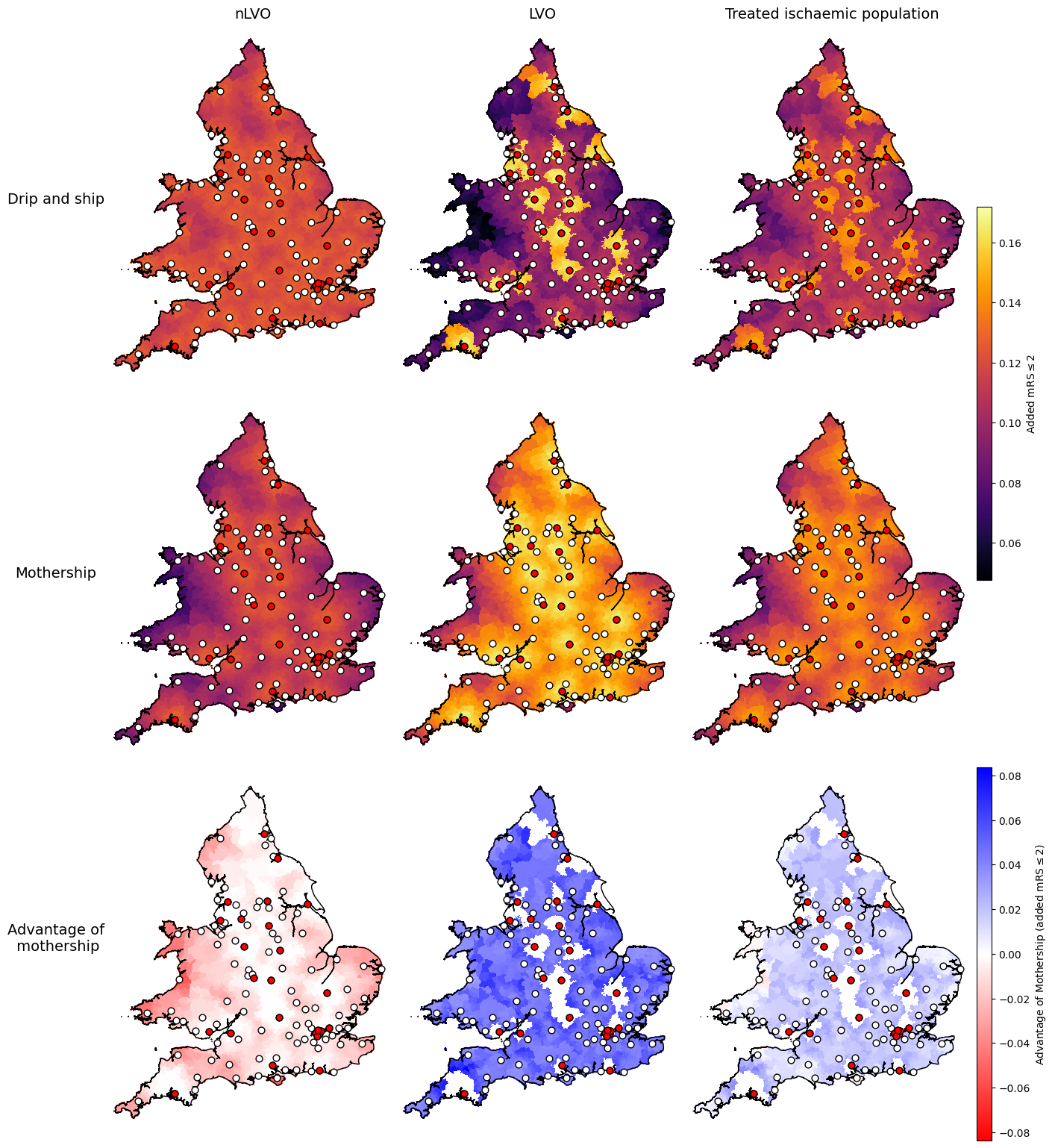
Settings for proportion with mRS less than or equal to 2:
data_field = 'added_mrs_less_equal_2'
vmin_scenarios, vmax_scenarios = find_vlims_scenarios(lsoa_data_gdf, data_field, cohorts)
vmin_diff, vmax_diff = find_vlims_diff(lsoa_data_gdf, data_field, cohorts)
params_added_mrs_less_equal_2 = {
'shared': {
'vmin': vmin_scenarios,
'vmax': vmax_scenarios,
'cmap': 'inferno',
'cbar_label': r'Added mRS$\leq$2'
},
'diff': {
'vmin': vmin_diff,
'vmax': vmax_diff,
'cmap': 'bwr_r',
'cbar_label': r'Advantage of Mothership (added mRS$\leq$2)'
},
}
axs_cols_added_mrs_less_equal_2 = [
# First row:
[f'drip_ship_nlvo_ivt_{data_field}',
f'drip_ship_lvo_mix_{data_field}',
f'drip_ship_weighted_treated_{data_field}'],
# Second row:
[f'mothership_nlvo_ivt_{data_field}',
f'mothership_lvo_mix_{data_field}',
f'mothership_weighted_treated_{data_field}'],
# Third row:
[f'nlvo_ivt_{data_field}_mothership_minus_dripship',
f'lvo_mix_{data_field}_mothership_minus_dripship',
f'weighted_treated_{data_field}_mothership_minus_dripship'],
]
plot_nine(
lsoa_data_gdf,
axs_cols_added_mrs_less_equal_2,
axs_params,
params_added_mrs_less_equal_2,
hospitals_gdf,
outline,
col_titles,
row_titles,
savename=os.path.join(dir_output, f'{data_field}_nine_in_one.jpg')
)
Conclusion#
The maps show similar results for the three outcome types: added utility, mean shift in mRS, and proportion with mRS score of 2 or less.
The advantage of mothership is worse for nLVO patients (the map is mostly red), better for LVO patients (the map is mostly blue), and generally slightly better overall for this mix of treated ischaemic patients.
Some parts of the combined treated ischaemic maps break the trend and have worse outcomes with mothership (drawn in red). The easiest to spot on this map is half-way up the west coast of Wales near Aberystwyth. The following notebook digs into these outlier areas more to see why they break the trend.