National mRS distributions#
This notebook creates a plot of the mRS distributions averaged across England and Wales.
Plain English summary#
This notebook loads the data that we created in a previous notebook. It loads the mRS distributions averaged across all parts of England and Wales where redirection from the nearest stroke unit to an MT centre makes a difference to which stroke unit is attended first.
Aims#
Plot the national mRS distributions for these cohorts:
nLVO with IVT
LVO with mixed treatments
treated ischaemic population
Methods#
Load in the existing data.
Create new no-treatment mRS distributions for the weighted combination by combining the separate nLVO and LVO no-treatment distributions in the required proportions.
Plot the mRS distributions and the cumulative probabilities.
dir_output = 'output'
limit_to_england = False
Import packages#
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import os
import stroke_outcome.outcome_utilities
pd.set_option('display.max_rows', 150)
stroke_outcome.__version__
'0.1.6'
Load data#
Results from outcome model:
df_mrs_national_cumsum = pd.read_csv(os.path.join(dir_output, 'cohort_mrs_dists_weighted_national.csv'), index_col=0)
df_mrs_national_cumsum
| mRS<=0 | mRS<=1 | mRS<=2 | mRS<=3 | mRS<=4 | mRS<=5 | mRS<=6 | |
|---|---|---|---|---|---|---|---|
| drip_ship_lvo_ivt | 0.10356 | 0.19544 | 0.32788 | 0.49012 | 0.70857 | 0.82316 | 1.0 |
| drip_ship_lvo_ivt_mt | 0.11472 | 0.22605 | 0.37176 | 0.54453 | 0.75047 | 0.85084 | 1.0 |
| drip_ship_lvo_mix | 0.11382 | 0.22365 | 0.36835 | 0.54031 | 0.74723 | 0.84869 | 1.0 |
| drip_ship_lvo_mt | 0.11449 | 0.22589 | 0.37170 | 0.54455 | 0.75052 | 0.85086 | 1.0 |
| drip_ship_nlvo_ivt | 0.36134 | 0.58906 | 0.70508 | 0.82416 | 0.92123 | 0.95515 | 1.0 |
| drip_ship_weighted | 0.18250 | 0.39015 | 0.51547 | 0.65399 | 0.81988 | 0.89575 | 1.0 |
| drip_ship_weighted_treated | 0.23655 | 0.40482 | 0.53531 | 0.68105 | 0.83350 | 0.90148 | 1.0 |
| mothership_lvo_ivt | 0.09705 | 0.18809 | 0.32101 | 0.48329 | 0.70384 | 0.81982 | 1.0 |
| mothership_lvo_ivt_mt | 0.15523 | 0.27670 | 0.42080 | 0.59378 | 0.78327 | 0.87247 | 1.0 |
| mothership_lvo_mix | 0.15072 | 0.26984 | 0.41306 | 0.58522 | 0.77711 | 0.86839 | 1.0 |
| mothership_lvo_mt | 0.15523 | 0.27670 | 0.42080 | 0.59378 | 0.78327 | 0.87247 | 1.0 |
| mothership_nlvo_ivt | 0.34442 | 0.57736 | 0.69431 | 0.81494 | 0.91617 | 0.95206 | 1.0 |
| mothership_weighted | 0.18504 | 0.39450 | 0.51975 | 0.65849 | 0.82301 | 0.89784 | 1.0 |
| mothership_weighted_treated | 0.24676 | 0.42231 | 0.55251 | 0.69911 | 0.84606 | 0.90988 | 1.0 |
df_mrs_national_noncum = pd.read_csv(os.path.join(dir_output, 'cohort_mrs_dists_weighted_national_noncum.csv'), index_col=0)
df_mrs_national_noncum
| mRS=0 | mRS=1 | mRS=2 | mRS=3 | mRS=4 | mRS=5 | mRS=6 | |
|---|---|---|---|---|---|---|---|
| drip_ship_lvo_ivt | 0.10356 | 0.09188 | 0.13244 | 0.16224 | 0.21845 | 0.11459 | 0.17684 |
| drip_ship_lvo_ivt_mt | 0.11472 | 0.11133 | 0.14571 | 0.17277 | 0.20594 | 0.10037 | 0.14916 |
| drip_ship_lvo_mix | 0.11382 | 0.10983 | 0.14470 | 0.17197 | 0.20692 | 0.10146 | 0.15131 |
| drip_ship_lvo_mt | 0.11449 | 0.11139 | 0.14582 | 0.17284 | 0.20597 | 0.10034 | 0.14914 |
| drip_ship_nlvo_ivt | 0.36134 | 0.22771 | 0.11603 | 0.11907 | 0.09707 | 0.03393 | 0.04485 |
| drip_ship_weighted | 0.18250 | 0.20765 | 0.12532 | 0.13852 | 0.16589 | 0.07586 | 0.10425 |
| drip_ship_weighted_treated | 0.23655 | 0.16828 | 0.13048 | 0.14574 | 0.15245 | 0.06798 | 0.09852 |
| mothership_lvo_ivt | 0.09705 | 0.09104 | 0.13293 | 0.16228 | 0.22054 | 0.11599 | 0.18018 |
| mothership_lvo_ivt_mt | 0.15523 | 0.12147 | 0.14409 | 0.17298 | 0.18949 | 0.08921 | 0.12753 |
| mothership_lvo_mix | 0.15072 | 0.11911 | 0.14323 | 0.17215 | 0.19189 | 0.09128 | 0.13161 |
| mothership_lvo_mt | 0.15523 | 0.12147 | 0.14409 | 0.17298 | 0.18949 | 0.08921 | 0.12753 |
| mothership_nlvo_ivt | 0.34442 | 0.23294 | 0.11695 | 0.12062 | 0.10124 | 0.03589 | 0.04794 |
| mothership_weighted | 0.18504 | 0.20946 | 0.12525 | 0.13873 | 0.16452 | 0.07483 | 0.10216 |
| mothership_weighted_treated | 0.24676 | 0.17555 | 0.13020 | 0.14660 | 0.14695 | 0.06382 | 0.09012 |
df_mrs_national_std = pd.read_csv(os.path.join(dir_output, 'cohort_mrs_dists_weighted_national_std.csv'), index_col=0)
df_mrs_national_std
| mRS=0 | mRS=1 | mRS=2 | mRS=3 | mRS=4 | mRS=5 | mRS=6 | |
|---|---|---|---|---|---|---|---|
| drip_ship_lvo_ivt | 0.00262 | 0.00029 | 0.00022 | 0.00003 | 0.00083 | 0.00055 | 0.00129 |
| drip_ship_lvo_ivt_mt | 0.00983 | 0.00399 | 0.00143 | 0.00129 | 0.00452 | 0.00355 | 0.00729 |
| drip_ship_lvo_mix | 0.00784 | 0.00317 | 0.00112 | 0.00102 | 0.00360 | 0.00283 | 0.00580 |
| drip_ship_lvo_mt | 0.01037 | 0.00373 | 0.00054 | 0.00080 | 0.00460 | 0.00347 | 0.00723 |
| drip_ship_nlvo_ivt | 0.00667 | 0.00215 | 0.00039 | 0.00064 | 0.00160 | 0.00074 | 0.00116 |
| drip_ship_weighted | 0.00128 | 0.00048 | 0.00015 | 0.00015 | 0.00049 | 0.00037 | 0.00074 |
| drip_ship_weighted_treated | 0.00515 | 0.00192 | 0.00060 | 0.00060 | 0.00198 | 0.00147 | 0.00298 |
| mothership_lvo_ivt | 0.00568 | 0.00090 | 0.00035 | 0.00004 | 0.00188 | 0.00128 | 0.00309 |
| mothership_lvo_ivt_mt | 0.01227 | 0.00250 | 0.00093 | 0.00032 | 0.00475 | 0.00307 | 0.00577 |
| mothership_lvo_mix | 0.00978 | 0.00199 | 0.00074 | 0.00025 | 0.00378 | 0.00245 | 0.00460 |
| mothership_lvo_mt | 0.01227 | 0.00250 | 0.00093 | 0.00032 | 0.00475 | 0.00307 | 0.00577 |
| mothership_nlvo_ivt | 0.01520 | 0.00444 | 0.00075 | 0.00131 | 0.00387 | 0.00187 | 0.00297 |
| mothership_weighted | 0.00224 | 0.00060 | 0.00013 | 0.00016 | 0.00067 | 0.00038 | 0.00068 |
| mothership_weighted_treated | 0.00901 | 0.00242 | 0.00053 | 0.00066 | 0.00271 | 0.00154 | 0.00275 |
cols_each_scen = df_mrs_national_noncum.index.values
cols_each_scen
array(['drip_ship_lvo_ivt', 'drip_ship_lvo_ivt_mt', 'drip_ship_lvo_mix',
'drip_ship_lvo_mt', 'drip_ship_nlvo_ivt', 'drip_ship_weighted',
'drip_ship_weighted_treated', 'mothership_lvo_ivt',
'mothership_lvo_ivt_mt', 'mothership_lvo_mix', 'mothership_lvo_mt',
'mothership_nlvo_ivt', 'mothership_weighted',
'mothership_weighted_treated'], dtype=object)
Reference no-treatment mRS distributions:
mrs_dists, mrs_dists_notes = (
stroke_outcome.outcome_utilities.import_mrs_dists_from_file())
mrs_dists
| mRS<=0 | mRS<=1 | mRS<=2 | mRS<=3 | mRS<=4 | mRS<=5 | mRS<=6 | |
|---|---|---|---|---|---|---|---|
| Stroke type | |||||||
| pre_stroke_nlvo | 0.583 | 0.746 | 0.850 | 0.951 | 0.993 | 1.000 | 1 |
| pre_stroke_lvo | 0.408 | 0.552 | 0.672 | 0.838 | 0.956 | 1.000 | 1 |
| no_treatment_lvo | 0.050 | 0.129 | 0.265 | 0.429 | 0.676 | 0.811 | 1 |
| no_treatment_nlvo | 0.198 | 0.460 | 0.580 | 0.708 | 0.856 | 0.918 | 1 |
| no_effect_nlvo_ivt_deaths | 0.196 | 0.455 | 0.574 | 0.701 | 0.847 | 0.908 | 1 |
| no_effect_lvo_ivt_deaths | 0.048 | 0.124 | 0.255 | 0.414 | 0.653 | 0.783 | 1 |
| no_effect_lvo_mt_deaths | 0.048 | 0.124 | 0.255 | 0.412 | 0.649 | 0.779 | 1 |
| t0_treatment_nlvo_ivt | 0.445 | 0.642 | 0.752 | 0.862 | 0.941 | 0.967 | 1 |
| t0_treatment_lvo_ivt | 0.140 | 0.233 | 0.361 | 0.522 | 0.730 | 0.838 | 1 |
| t0_treatment_lvo_mt | 0.306 | 0.429 | 0.548 | 0.707 | 0.851 | 0.915 | 1 |
mrs_dist_nlvo_no_treatment = mrs_dists.loc['no_treatment_nlvo'].values
mrs_dist_lvo_no_treatment = mrs_dists.loc['no_treatment_lvo'].values
mrs_dist_nlvo_no_treatment_noncum = np.diff(mrs_dist_nlvo_no_treatment, prepend=0.0)
mrs_dist_lvo_no_treatment_noncum = np.diff(mrs_dist_lvo_no_treatment, prepend=0.0)
Stroke type proportions:
proportions = pd.read_csv(
os.path.join(dir_output, 'patient_proportions.csv'),
index_col=0, header=None).squeeze()
proportions
0
haemorrhagic 0.13600
lvo_no_treatment 0.14648
lvo_ivt_only 0.00840
lvo_ivt_mt 0.08500
lvo_mt_only 0.01500
nlvo_no_treatment 0.50252
nlvo_ivt 0.10660
Name: 1, dtype: float64
Calculate weighted no treatment dist:#
prop_lvo_treated = 0.0
prop_nlvo_treated = 0.0
for key, value in proportions.items():
if ('nlvo' in key) & ('no_treatment' not in key):
prop_nlvo_treated += value
elif ('lvo' in key) & ('no_treatment' not in key):
prop_lvo_treated += value
prop_lvo_treated, prop_nlvo_treated
(0.10840000000000001, 0.1066)
prop_lvo_treated / (prop_nlvo_treated + prop_lvo_treated), prop_nlvo_treated / (prop_nlvo_treated + prop_lvo_treated)
(0.5041860465116279, 0.49581395348837204)
mrs_dist_weighted_no_treatment_noncum = (
(mrs_dist_nlvo_no_treatment_noncum * prop_nlvo_treated) +
(mrs_dist_lvo_no_treatment_noncum * prop_lvo_treated)
)
# Remove the non-treated patients:
mrs_dist_weighted_no_treatment_noncum = (
mrs_dist_weighted_no_treatment_noncum / (prop_nlvo_treated + prop_lvo_treated)
)
np.sum(mrs_dist_weighted_no_treatment_noncum)
1.0
# Same structure as the other distribution dictionaries:
dict_no_treat = {
'nlvo': {
'values': mrs_dist_nlvo_no_treatment_noncum,
'cumsum': np.cumsum(mrs_dist_nlvo_no_treatment_noncum)
},
'lvo': {
'values': mrs_dist_lvo_no_treatment_noncum,
'cumsum': np.cumsum(mrs_dist_lvo_no_treatment_noncum)
},
# 'weighted': {
# 'values': mrs_dist_weighted_no_treatment_noncum,
# 'cumsum': np.cumsum(mrs_dist_weighted_no_treatment_noncum)
# },
'weighted_treated': {
'values': mrs_dist_weighted_no_treatment_noncum,
'cumsum': np.cumsum(mrs_dist_weighted_no_treatment_noncum)
},
}
Gather data for plots#
def gather_dists(scenario, df_mrs_national_noncum, df_mrs_national_std, df_mrs_national_cumsum):
d = {}
for cohort in ['nlvo_ivt', 'lvo_mix', 'weighted_treated']:
ind = f'{scenario}_{cohort}'
d[cohort] = {}
d[cohort]['values'] = df_mrs_national_noncum.loc[ind]
d[cohort]['std'] = df_mrs_national_std.loc[ind]
d[cohort]['cumsum'] = df_mrs_national_cumsum.loc[ind]
return d
dict_drip_ship = gather_dists('drip_ship', df_mrs_national_noncum, df_mrs_national_std, df_mrs_national_cumsum)
dict_mothership = gather_dists('mothership', df_mrs_national_noncum, df_mrs_national_std, df_mrs_national_cumsum)
Plot#
colour_drip = '#00517f' # seaborn colorblind blue darker
colour_moth = '#0072b2' # seaborn colorblind blue
colour_no_treat = 'DarkGray'
fig, axs = plt.subplots(2, 3, figsize=(15, 7), gridspec_kw={'wspace': 0.1})
bar_width = 0.25
offsets = [-bar_width, 0.0, bar_width]
legend_drawn = False
for c, cohort in enumerate(['nlvo_ivt', 'lvo_mix', 'weighted_treated']):
dist_drip = dict_drip_ship[cohort]['values']
std_drip = dict_drip_ship[cohort]['std']
dist_cumsum_drip = dict_drip_ship[cohort]['cumsum']
dist_moth = dict_mothership[cohort]['values']
std_moth = dict_mothership[cohort]['std']
dist_cumsum_moth = dict_mothership[cohort]['cumsum']
if 'nlvo' in cohort:
ax = axs[0, 0]
ax_cumsum = axs[1, 0]
dist_no_treat = dict_no_treat['nlvo']['values']
dist_cumsum_no_treat = dict_no_treat['nlvo']['cumsum']
elif 'lvo' in cohort:
ax = axs[0, 1]
ax_cumsum = axs[1, 1]
dist_no_treat = dict_no_treat['lvo']['values']
dist_cumsum_no_treat = dict_no_treat['lvo']['cumsum']
elif 'treated' in cohort:
ax = axs[0, 2]
ax_cumsum = axs[1, 2]
dist_no_treat = dict_no_treat['weighted_treated']['values']
dist_cumsum_no_treat = dict_no_treat['weighted_treated']['cumsum']
# else:
# ax = axs[0, -1]
# ax_cumsum = axs[1, -1]
# dist_no_treat = dict_no_treat['weighted']['values']
# dist_cumsum_no_treat = dict_no_treat['weighted']['cumsum']
ax.bar(
np.arange(7) + offsets[0], dist_drip, yerr=std_drip, capsize=2.0,
facecolor=colour_drip, label='Drip & ship', width=bar_width
)
ax.bar(
np.arange(7) + offsets[1], dist_moth, yerr=std_moth, capsize=2.0,
facecolor=colour_moth, label='Mothership', width=bar_width
)
ax.bar(
np.arange(7) + offsets[2], dist_no_treat,
facecolor=colour_no_treat, label='No treatment', width=bar_width
)
ax_cumsum.plot(np.arange(7), dist_cumsum_drip, color=colour_drip, linestyle='-', label='Drip & ship')
ax_cumsum.plot(np.arange(7), dist_cumsum_moth, color=colour_moth, linestyle='--', label='Mothership')
ax_cumsum.plot(np.arange(7), dist_cumsum_no_treat, color=colour_no_treat, linestyle=':', label='No treatment')
if c == 2:
ax.legend(bbox_to_anchor=(1.0, 0.5), loc='center left')
ax_cumsum.legend(bbox_to_anchor=(1.0, 0.5), loc='center left')
legend_drawn = True
for i, ax_list in enumerate(axs):
ylim = [0, 0.4] if i == 0 else [0.0, 1.05]
for j, ax in enumerate(ax_list):
ax.set_ylim(*ylim)
ax.set_xticks(range(7))
if (j > 0): # & (j < len(ax_list) - 1):
ax.set_yticklabels([])
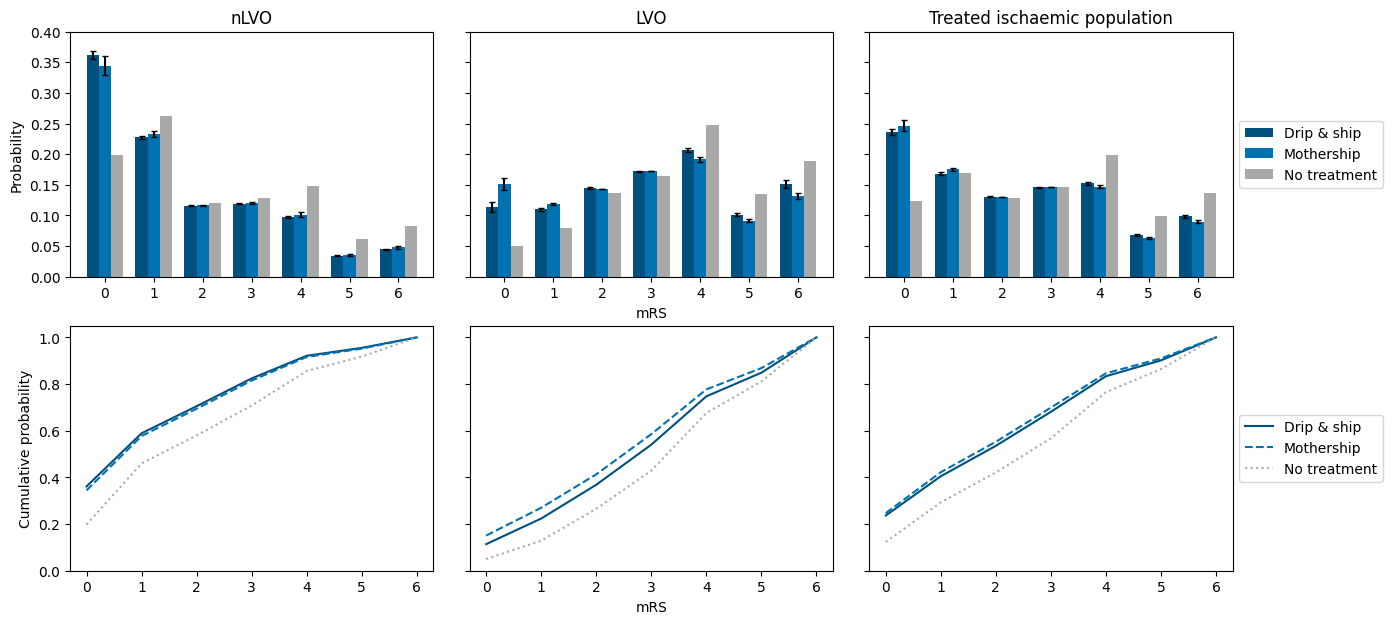
axs[0, 0].set_title('nLVO')
axs[0, 1].set_title('LVO')
axs[0, 2].set_title('Treated ischaemic population')
# axs[0, -1].set_title('Total\nischaemic population')
axs[0, 0].set_ylabel('Probability')
# axs[0, -1].set_ylabel('Probability')
# axs[0, -1].yaxis.set_tick_params(labelleft=True) # turn the tick labels back on
axs[0, 1].set_xlabel('mRS')
# axs[0, -1].set_xlabel('mRS')
axs[1, 0].set_ylabel('Cumulative probability')
# axs[1, -1].set_ylabel('Cumulative probability')
# axs[1, -1].yaxis.set_tick_params(labelleft=True) # turn the tick labels back on
axs[1, 1].set_xlabel('mRS')
# axs[1, -1].set_xlabel('mRS')
# axs[0, 3].axis('off')
# axs[1, 3].axis('off')
plt.savefig(os.path.join(dir_output, 'mrs_dists_redirection_areas.png'), bbox_inches='tight')
plt.show()
Find mean values#
Import utility scores
utility_dists, utility_dists_notes = (
stroke_outcome.outcome_utilities.import_utility_dists_from_file())
utility_weights = utility_dists.loc['Wang2020'].values
dist_dicts = {
'drip_ship': dict_drip_ship,
'mothership': dict_mothership,
'no_treat': dict_no_treat
}
df_means = pd.DataFrame()
for scenario, dist_dict in dist_dicts.items():
for cohort, d_dict in dist_dict.items():
if 'nlvo' in cohort:
dist_no_treat = dict_no_treat['nlvo']['values']
elif 'lvo' in cohort:
dist_no_treat = dict_no_treat['lvo']['values']
elif 'treated' in cohort:
dist_no_treat = dict_no_treat['weighted_treated']['values']
else:
print('help')
# dist_no_treat = dict_no_treat['weighted']['values']
mean_mrs_no_treat = np.mean(dist_no_treat * np.arange(7))
mean_util_no_treat = np.mean(dist_no_treat * utility_weights)
mrsleq2_no_treat = np.sum(dist_no_treat[:3])
dist = d_dict['values']
ind = f'{scenario}_{cohort}'
df_means.loc[ind, 'mean_mrs'] = np.mean(dist * np.arange(7))
df_means.loc[ind, 'mean_util'] = np.mean(dist * utility_weights)
df_means.loc[ind, 'mrsleq2'] = np.sum(dist[:3])
df_means.loc[ind, 'mean_mrs_shift'] = (
df_means.loc[ind, 'mean_mrs'] - mean_mrs_no_treat)
df_means.loc[ind, 'mean_util_shift'] = (
df_means.loc[ind, 'mean_util'] - mean_util_no_treat)
df_means.loc[ind, 'mrsleq2_shift'] = (
df_means.loc[ind, 'mrsleq2'] - mrsleq2_no_treat)
df_means
| mean_mrs | mean_util | mrsleq2 | mean_mrs_shift | mean_util_shift | mrsleq2_shift | |
|---|---|---|---|---|---|---|
| drip_ship_nlvo_ivt | 0.234859 | 0.102172 | 0.705080 | -0.090856 | 0.016509 | 0.125080 |
| drip_ship_lvo_mix | 0.451140 | 0.061546 | 0.368350 | -0.068860 | 0.014031 | 0.103350 |
| drip_ship_weighted_treated | 0.343897 | 0.081689 | 0.535310 | -0.079773 | 0.015260 | 0.114129 |
| mothership_nlvo_ivt | 0.242964 | 0.100770 | 0.694310 | -0.082750 | 0.015107 | 0.114310 |
| mothership_lvo_mix | 0.419377 | 0.067532 | 0.413060 | -0.100623 | 0.020016 | 0.148060 |
| mothership_weighted_treated | 0.331910 | 0.084012 | 0.552510 | -0.091760 | 0.017582 | 0.131329 |
| no_treat_nlvo | 0.325714 | 0.085663 | 0.580000 | 0.000000 | 0.000000 | 0.000000 |
| no_treat_lvo | 0.520000 | 0.047516 | 0.265000 | 0.000000 | 0.000000 | 0.000000 |
| no_treat_weighted_treated | 0.423670 | 0.066430 | 0.421181 | 0.000000 | 0.000000 | 0.000000 |
Save a copy of data behind the figure#
Keep a copy of the separate LVO cohorts even though the figure only shows the combined LVO cohort.
Add the no-treatment distributions into the starting data:
for key, value in dict_no_treat.items():
ind = f'no_treatment_{key}'
df_mrs_national_noncum.loc[ind] = value
df_mrs_national_std.loc[ind] = pd.NA
# Save to file:
df_mrs_national_noncum.to_csv(os.path.join(dir_output, 'fig_data_mrs_dists.csv'))
df_mrs_national_std.to_csv(os.path.join(dir_output, 'fig_data_mrs_dists_std.csv'))
df_means.to_csv(os.path.join(dir_output, 'fig_data_mrs_dists_means.csv'))
Conclusions#
For nLVO, the mRS distributions are weighted slightly lower (less disability) for the drip-and-ship scenario. For LVO, they are weighted lower (less disability) for the mothership scenario. For the combined cohort, the mothership scenario just wins out. This is most easily seen by looking at which scenario is highest on the cumulative probability plots.