Descriptive analysis of stroke pathway data
Contents
Descriptive analysis of stroke pathway data#
Aim#
Describe key pathway statistics
Restrict data to stroke teams with at least 300 admissions (over three years) and 10 patients receiving thrombolysis.
Analyse pathway statistics by stroke team, and combined across all stroke teams.
Repeat team and combined analysis by age category (under aged 80 years, and aged 80+).
Create histograms of variation in key statistics across stroke teams.
Analyse onset-to-arrival (and breakdown by mild and non-mild stroke)
Import libraries and data#
Data has been restricted to stroke teams with at least 300 admissions, with at least 10 patients receiving thrombolysis, over three years.
# import libraries
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
# import data
raw_data = pd.read_csv(
'./../data/2019-11-04-HQIP303-Exeter_MA.csv', low_memory=False)
headings = list(raw_data)
print (headings)
['StrokeTeam', 'PatientUID', 'Pathway', 'S1AgeOnArrival', 'MoreEqual80y', 'S1Gender', 'S1Ethnicity', 'S1OnsetInHospital', 'S1OnsetToArrival_min', 'S1OnsetDateType', 'S1OnsetTimeType', 'S1ArriveByAmbulance', 'S1AdmissionHour', 'S1AdmissionDay', 'S1AdmissionQuarter', 'S1AdmissionYear', 'CongestiveHeartFailure', 'Hypertension', 'AtrialFibrillation', 'Diabetes', 'StrokeTIA', 'AFAntiplatelet', 'AFAnticoagulent', 'AFAnticoagulentVitK', 'AFAnticoagulentDOAC', 'AFAnticoagulentHeparin', 'S2INR', 'S2INRHigh', 'S2INRNK', 'S2NewAFDiagnosis', 'S2RankinBeforeStroke', 'Loc', 'LocQuestions', 'LocCommands', 'BestGaze', 'Visual', 'FacialPalsy', 'MotorArmLeft', 'MotorArmRight', 'MotorLegLeft', 'MotorLegRight', 'LimbAtaxia', 'Sensory', 'BestLanguage', 'Dysarthria', 'ExtinctionInattention', 'S2NihssArrival', 'S2BrainImagingTime_min', 'S2StrokeType', 'S2Thrombolysis', 'Haemorrhagic', 'TimeWindow', 'Comorbidity', 'Medication', 'Refusal', 'Age', 'Improving', 'TooMildSevere', 'TimeUnknownWakeUp', 'OtherMedical', 'S2ThrombolysisTime_min', 'S2TIAInLastMonth']
Show data types
raw_data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 246676 entries, 0 to 246675
Data columns (total 62 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 StrokeTeam 246676 non-null object
1 PatientUID 246676 non-null object
2 Pathway 246676 non-null int64
3 S1AgeOnArrival 246676 non-null object
4 MoreEqual80y 246676 non-null object
5 S1Gender 246676 non-null object
6 S1Ethnicity 246676 non-null object
7 S1OnsetInHospital 246676 non-null object
8 S1OnsetToArrival_min 154640 non-null float64
9 S1OnsetDateType 246676 non-null object
10 S1OnsetTimeType 246676 non-null object
11 S1ArriveByAmbulance 232503 non-null object
12 S1AdmissionHour 246676 non-null object
13 S1AdmissionDay 246676 non-null object
14 S1AdmissionQuarter 246676 non-null object
15 S1AdmissionYear 246676 non-null int64
16 CongestiveHeartFailure 246676 non-null object
17 Hypertension 246676 non-null object
18 AtrialFibrillation 246676 non-null object
19 Diabetes 246676 non-null object
20 StrokeTIA 246676 non-null object
21 AFAntiplatelet 47691 non-null object
22 AFAnticoagulent 120409 non-null object
23 AFAnticoagulentVitK 93603 non-null object
24 AFAnticoagulentDOAC 93603 non-null object
25 AFAnticoagulentHeparin 93603 non-null object
26 S2INR 4240 non-null float64
27 S2INRHigh 93603 non-null object
28 S2INRNK 93603 non-null object
29 S2NewAFDiagnosis 70588 non-null object
30 S2RankinBeforeStroke 246676 non-null int64
31 Loc 246676 non-null int64
32 LocQuestions 236305 non-null float64
33 LocCommands 236579 non-null float64
34 BestGaze 235003 non-null float64
35 Visual 233231 non-null float64
36 FacialPalsy 236381 non-null float64
37 MotorArmLeft 236475 non-null float64
38 MotorArmRight 236478 non-null float64
39 MotorLegLeft 236205 non-null float64
40 MotorLegRight 236202 non-null float64
41 LimbAtaxia 232480 non-null float64
42 Sensory 233464 non-null float64
43 BestLanguage 236047 non-null float64
44 Dysarthria 235035 non-null float64
45 ExtinctionInattention 234707 non-null float64
46 S2NihssArrival 228279 non-null float64
47 S2BrainImagingTime_min 245535 non-null float64
48 S2StrokeType 245535 non-null object
49 S2Thrombolysis 246676 non-null object
50 Haemorrhagic 215844 non-null object
51 TimeWindow 215844 non-null object
52 Comorbidity 215844 non-null object
53 Medication 215844 non-null object
54 Refusal 215844 non-null object
55 Age 215844 non-null object
56 Improving 215844 non-null object
57 TooMildSevere 215844 non-null object
58 TimeUnknownWakeUp 215844 non-null object
59 OtherMedical 215844 non-null object
60 S2ThrombolysisTime_min 28482 non-null float64
61 S2TIAInLastMonth 21151 non-null object
dtypes: float64(19), int64(4), object(39)
memory usage: 116.7+ MB
Average yearly admissions#
int(raw_data.shape[0]/3)
82225
Restrict original data to hospitals with at least 300 admissions + 10 thrombolysis patients#
keep = []
groups = raw_data.groupby('StrokeTeam') # creates a new object of groups of data
for index, group_df in groups: # each group has an index and a dataframe of data
# Skip if total admissions less than 300 or total thrombolysis < 10
admissions = group_df.shape[0]
thrombolysis_received = group_df['S2Thrombolysis'] == 'Yes'
if (admissions < 300) or (thrombolysis_received.sum() < 10):
continue
else:
keep.append(group_df)
# Concatenate output
data = pd.DataFrame()
data = pd.concat(keep)
print (data.shape)
(239505, 62)
Stroke types#
data['S2StrokeType'].value_counts()/data['S2StrokeType'].shape[0]
Infarction 0.873435
Primary Intracerebral Haemorrhage 0.122106
Name: S2StrokeType, dtype: float64
Compare thrombolysis use for out-of-hospital and in-hospital stroke onset#
# Create boolean feature for use of thrombolysis
data['use_thrombolysis'] = data['S2Thrombolysis'] == 'Yes'
# Show percentage of strokes have in hospital onset
in_hospital_onset = data['S1OnsetInHospital'] == 'Yes'
percent_in_hosp = in_hospital_onset.mean() * 100
print (
f'Percentage of stroke with in-hospital onset: {percent_in_hosp:0.1f}')
# In hospital onset
mask = data['S1OnsetInHospital'] == 'Yes'
thrombolysis_in = data[mask]['use_thrombolysis'].mean() * 100
print (
f'Thrombolysis use (%) for in-hospital-onset {thrombolysis_in:0.1f}')
# Out of hospital onset
mask = data['S1OnsetInHospital'] == 'No'
thrombolysis_out = data[mask]['use_thrombolysis'].mean() * 100
print (
f'Thrombolysis use (%) for out-of-hospital-onset {thrombolysis_out:0.1f}')
Percentage of stroke with in-hospital onset: 5.3
Thrombolysis use (%) for in-hospital-onset 12.3
Thrombolysis use (%) for out-of-hospital-onset 11.8
Restrict data to out-of-hospital onset#
mask = data['S1OnsetInHospital'] == 'No'
data = data[mask]
Function for analysis by stroke team#
def analyse_by_team(input_data):
# Copy data
data = input_data.copy()
# Set up results lists
stroke_team = []
admissions = []
age_80_plus = []
onset_known = []
rankin_before_stroke_all = []
nihss_arrival_all = []
known_arrival_within_4hrs = []
rankin_before_stroke_4hrs = []
nihss_arrival_4hrs = []
onset_arrival_mins_mu = []
onset_arrival_mins_sigma = []
scan_within_4_hrs = []
arrival_scan_mins_mu = []
arrival_scan_mins_sigma = []
scan_needle_mins_mu = []
scan_needle_mins_sigma = []
arrival_needle_mins_mu = []
arrival_needle_mins_sigma = []
onset_needle_mins_mu = []
onset_needle_mins_sigma = []
thrombolysis_rate = []
thrombolysis_after_180_min = []
thrombolysis_after_270_min = []
eligible = []
# Split data by stroke team
groups = data.groupby('StrokeTeam') # creates a new object of groups of data
group_count = 0
for index, group_df in groups:
group_count += 1
# Record stroke team
stroke_team.append(index)
# Record admission numbers
admissions.append(group_df.shape[0])
# Get thrombolysis rate
thrombolysed = group_df['S2Thrombolysis'] == 'Yes'
thrombolysis_rate.append(thrombolysed.mean())
# Get Rankin before stroke and NIHSS on arrival
rankin_before_stroke_all.append(group_df['S2RankinBeforeStroke'].mean())
nihss_arrival_all.append(group_df['S2NihssArrival'].mean())
# Record onset known proportion and remove rest
f = lambda x: x in ['Precise', 'Best estimate']
mask = group_df['S1OnsetTimeType'].apply(f)
onset_known.append(mask.mean())
group_df = group_df[mask]
# Record onset <4 hours and remove rest
mask = group_df['S1OnsetToArrival_min'] <= 240
known_arrival_within_4hrs.append(mask.mean())
group_df = group_df[mask]
# Get Rankin before stroke and NIHSS on arrival
rankin_before_stroke_4hrs.append(
group_df['S2RankinBeforeStroke'].mean())
nihss_arrival_4hrs.append(group_df['S2NihssArrival'].mean())
# Calc proportion 80+ (of those arriving within 4 hours)
age_filter = group_df['MoreEqual80y'] == 'Yes'
age_80_plus.append(age_filter.mean())
# Get onset to arrival (of those arriving within 4 hours)
onset_to_arrival = group_df['S1OnsetToArrival_min']
onset_arrival_mins_mu.append(onset_to_arrival.mean())
onset_arrival_mins_sigma.append(onset_to_arrival.std())
# Record scan within 4 hours of arrival (and remove the rest)
mask = group_df['S2BrainImagingTime_min'] <= 240
scan_within_4_hrs.append(mask.mean())
group_df = group_df[mask]
# Record arrival to scan (of those arriving within 4 hours)
arrival_to_scan = group_df['S2BrainImagingTime_min']
arrival_scan_mins_mu.append(arrival_to_scan.mean())
arrival_scan_mins_sigma.append(arrival_to_scan.std())
# Thrombolysis given (of those scanned within 4 hours of arrival)
thrombolysed_mask = group_df['S2Thrombolysis'] == 'Yes'
eligible.append(thrombolysed_mask.mean())
# Scan to needle
thrombolysed = group_df[thrombolysed_mask]
scan_to_needle = (thrombolysed['S2ThrombolysisTime_min'] -
thrombolysed['S2BrainImagingTime_min'])
scan_needle_mins_mu.append(scan_to_needle.mean())
scan_needle_mins_sigma.append(scan_to_needle.std())
# Arrival to needle
arrival_needle_mins_mu.append(
thrombolysed['S2ThrombolysisTime_min'].mean())
arrival_needle_mins_sigma.append(
thrombolysed['S2ThrombolysisTime_min'].std())
# Onset to needle
onset_to_needle = (thrombolysed['S1OnsetToArrival_min'] +
thrombolysed['S2ThrombolysisTime_min'])
onset_needle_mins_mu.append(onset_to_needle.mean())
onset_needle_mins_sigma.append(onset_to_needle.std())
# Proportions of thrombolysis after 180 or 270 min
thrombolysis_after_180 = onset_to_needle > 180
thrombolysis_after_270 = onset_to_needle > 270
thrombolysis_after_180_min.append(thrombolysis_after_180.mean())
thrombolysis_after_270_min.append(thrombolysis_after_270.mean())
df = pd.DataFrame()
df['stroke_team'] = stroke_team
df['thrombolysis_rate'] = thrombolysis_rate
df['arrivals'] = admissions
df['80_plus'] = age_80_plus
df['rankin_before_stroke_all'] = rankin_before_stroke_all
df['nihss_arrival_all'] = nihss_arrival_all
df['onset_known'] = onset_known
df['known_arrival_within_4hrs'] = known_arrival_within_4hrs
df['known_arrival_within_4hrs_all'] = \
df['onset_known'] * df['known_arrival_within_4hrs']
df['rankin_before_stroke_4hrs'] = rankin_before_stroke_4hrs
df['nihss_arrival_4hrs'] = nihss_arrival_4hrs
df['onset_arrival_mins_mu'] = onset_arrival_mins_mu
df['onset_arrival_mins_sigma'] = onset_arrival_mins_sigma
df['scan_within_4_hrs'] = scan_within_4_hrs
df['arrival_scan_mins_mu'] = arrival_scan_mins_mu
df['arrival_scan_mins_sigma'] = arrival_scan_mins_sigma
df['eligable'] = eligible
df['scan_needle_mins_mu'] = scan_needle_mins_mu
df['scan_needle_mins_sigma'] = scan_needle_mins_sigma
df['arrival_needle_mins_mu'] = arrival_needle_mins_mu
df['arrival_needle_mins_sigma'] = arrival_needle_mins_sigma
df['onset_needle_mins_mu'] = onset_needle_mins_mu
df['onset_needle_mins_sigma'] = onset_needle_mins_sigma
df['thrombolysis_after_180_min'] = thrombolysis_after_180_min
df['thrombolysis_after_270_min'] = thrombolysis_after_270_min
return df
Create analysis by team and show summary#
analysis_by_team = analyse_by_team(data)
analysis_by_team.to_csv('output/full_analysis_by_team.csv')
summary_by_hospital = analysis_by_team.describe().T
summary_by_hospital
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| thrombolysis_rate | 132.0 | 0.114536 | 0.035449 | 0.015306 | 0.093239 | 0.109547 | 0.133314 | 0.243174 |
| arrivals | 132.0 | 1718.333333 | 832.249409 | 303.000000 | 1133.000000 | 1632.500000 | 2265.250000 | 6118.000000 |
| 80_plus | 132.0 | 0.425635 | 0.056416 | 0.292308 | 0.387089 | 0.427573 | 0.457896 | 0.576220 |
| rankin_before_stroke_all | 132.0 | 1.040435 | 0.258761 | 0.512531 | 0.843626 | 1.032913 | 1.202876 | 1.860384 |
| nihss_arrival_all | 132.0 | 7.393900 | 0.948271 | 4.543634 | 6.698829 | 7.404792 | 8.067855 | 10.277926 |
| onset_known | 132.0 | 0.665679 | 0.134064 | 0.344784 | 0.573593 | 0.641978 | 0.752881 | 0.988275 |
| known_arrival_within_4hrs | 132.0 | 0.600743 | 0.089733 | 0.257034 | 0.557876 | 0.608542 | 0.662325 | 0.813584 |
| known_arrival_within_4hrs_all | 132.0 | 0.392147 | 0.062162 | 0.222646 | 0.352730 | 0.392458 | 0.430548 | 0.558317 |
| rankin_before_stroke_4hrs | 132.0 | 1.052002 | 0.264767 | 0.455108 | 0.877631 | 1.059910 | 1.204151 | 1.843863 |
| nihss_arrival_4hrs | 132.0 | 9.119220 | 1.107748 | 6.108696 | 8.419594 | 9.168358 | 9.916195 | 11.728546 |
| onset_arrival_mins_mu | 132.0 | 110.604942 | 6.663145 | 90.306383 | 106.872313 | 110.812667 | 115.087515 | 132.572368 |
| onset_arrival_mins_sigma | 132.0 | 52.401532 | 2.342917 | 46.456599 | 50.822598 | 52.119960 | 53.668951 | 63.899224 |
| scan_within_4_hrs | 132.0 | 0.947503 | 0.030163 | 0.847328 | 0.932828 | 0.953808 | 0.969191 | 1.000000 |
| arrival_scan_mins_mu | 132.0 | 42.991449 | 11.548507 | 18.635392 | 35.619539 | 42.907270 | 50.628576 | 92.923345 |
| arrival_scan_mins_sigma | 132.0 | 43.168750 | 6.917284 | 24.002919 | 38.927934 | 42.655178 | 47.546395 | 64.428643 |
| eligable | 132.0 | 0.301030 | 0.078284 | 0.073171 | 0.248810 | 0.293434 | 0.359811 | 0.497462 |
| scan_needle_mins_mu | 132.0 | 39.683068 | 12.020205 | 19.086768 | 30.691358 | 37.401892 | 45.949020 | 92.147059 |
| scan_needle_mins_sigma | 132.0 | 26.538991 | 8.507326 | 7.940403 | 21.293485 | 25.088262 | 29.942953 | 65.858529 |
| arrival_needle_mins_mu | 132.0 | 63.474563 | 15.121720 | 26.472222 | 54.488376 | 60.043478 | 71.720339 | 111.441558 |
| arrival_needle_mins_sigma | 132.0 | 32.814839 | 8.395361 | 8.195769 | 27.883198 | 32.228478 | 36.453541 | 69.820865 |
| onset_needle_mins_mu | 132.0 | 158.393399 | 13.147797 | 127.000000 | 149.722619 | 156.531821 | 166.692531 | 189.662338 |
| onset_needle_mins_sigma | 132.0 | 52.274870 | 5.907052 | 39.071523 | 48.582379 | 51.275398 | 54.895491 | 83.336167 |
| thrombolysis_after_180_min | 132.0 | 0.309629 | 0.093026 | 0.080000 | 0.250245 | 0.289664 | 0.356365 | 0.588235 |
| thrombolysis_after_270_min | 132.0 | 0.017742 | 0.015162 | 0.000000 | 0.008063 | 0.014728 | 0.025703 | 0.069853 |
# Save summary to csv
summary_by_hospital.to_csv('output/pathway_summary_by_hopsital.csv')
Analyse for all patients together#
# Set all StrokeTeam to same
single_group = data.copy()
single_group['StrokeTeam'] = "na"
analysis_all = analyse_by_team(single_group).T
# Add means to new DataFrame
stats_by_age = pd.DataFrame()
stats_by_age['all'] = analysis_all[0]
analysis_all.to_csv('output/pathway_summary_all.csv')
Analysis by under age 80 vs 80+#
Repeat general and hospital level analysis by age stratification
# Mask for under 80
mask = data['MoreEqual80y'] == 'No'
data_under_80 = data[mask]
# Mask for 80 plus
mask = data['MoreEqual80y'] == 'Yes'
data_80_plus = data[mask]
Under age 80#
analysis_by_team_under_80 = analyse_by_team(data_under_80)
summary_by_hospital_under_80 = analysis_by_team_under_80.describe().T
summary_by_hospital_under_80
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| thrombolysis_rate | 132.0 | 0.127182 | 0.038002 | 0.013064 | 0.104283 | 0.123472 | 0.150482 | 0.262436 |
| arrivals | 132.0 | 1003.037879 | 529.410379 | 173.000000 | 636.000000 | 941.500000 | 1286.500000 | 3999.000000 |
| 80_plus | 132.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| rankin_before_stroke_all | 132.0 | 0.642800 | 0.204014 | 0.221030 | 0.493693 | 0.609459 | 0.758040 | 1.420674 |
| nihss_arrival_all | 132.0 | 6.286699 | 0.856764 | 3.749164 | 5.766698 | 6.342502 | 6.799700 | 9.328110 |
| onset_known | 132.0 | 0.679982 | 0.130020 | 0.377171 | 0.588716 | 0.660804 | 0.760079 | 0.985591 |
| known_arrival_within_4hrs | 132.0 | 0.584347 | 0.090390 | 0.222084 | 0.537210 | 0.594057 | 0.649928 | 0.807645 |
| known_arrival_within_4hrs_all | 132.0 | 0.389793 | 0.060399 | 0.212589 | 0.351842 | 0.391457 | 0.423625 | 0.544338 |
| rankin_before_stroke_4hrs | 132.0 | 0.617723 | 0.214004 | 0.148148 | 0.462979 | 0.597158 | 0.752650 | 1.443884 |
| nihss_arrival_4hrs | 132.0 | 8.085814 | 1.080190 | 5.147959 | 7.459996 | 8.037950 | 8.714991 | 11.512821 |
| onset_arrival_mins_mu | 132.0 | 108.278108 | 6.901565 | 91.458333 | 103.999891 | 107.773886 | 113.655551 | 127.885057 |
| onset_arrival_mins_sigma | 132.0 | 52.352397 | 2.714954 | 46.708726 | 50.718741 | 52.128487 | 53.906830 | 61.943113 |
| scan_within_4_hrs | 132.0 | 0.949423 | 0.031731 | 0.833333 | 0.933499 | 0.958696 | 0.969423 | 1.000000 |
| arrival_scan_mins_mu | 132.0 | 42.717327 | 11.121473 | 18.496907 | 36.361292 | 42.133225 | 50.423320 | 94.846667 |
| arrival_scan_mins_sigma | 132.0 | 43.169589 | 7.199394 | 26.084190 | 38.407673 | 43.699258 | 47.659708 | 66.206585 |
| eligable | 132.0 | 0.334699 | 0.083152 | 0.073333 | 0.279830 | 0.333333 | 0.391261 | 0.524590 |
| scan_needle_mins_mu | 132.0 | 40.224581 | 12.107333 | 18.303333 | 32.081609 | 38.742367 | 46.597925 | 93.260870 |
| scan_needle_mins_sigma | 132.0 | 27.284895 | 10.235558 | 8.453550 | 21.021526 | 25.689491 | 28.915382 | 78.491646 |
| arrival_needle_mins_mu | 132.0 | 64.712453 | 15.445930 | 26.375000 | 54.799590 | 62.553309 | 73.987337 | 109.178571 |
| arrival_needle_mins_sigma | 132.0 | 34.256721 | 10.043992 | 8.365604 | 28.970198 | 32.771987 | 37.719027 | 83.959546 |
| onset_needle_mins_mu | 132.0 | 159.157281 | 13.620341 | 120.600000 | 149.626050 | 157.045433 | 168.504335 | 191.566667 |
| onset_needle_mins_sigma | 132.0 | 53.566155 | 7.108638 | 38.454895 | 49.181509 | 52.290158 | 56.504757 | 86.605961 |
| thrombolysis_after_180_min | 132.0 | 0.316502 | 0.094463 | 0.000000 | 0.252605 | 0.301239 | 0.375613 | 0.565217 |
| thrombolysis_after_270_min | 132.0 | 0.019459 | 0.018550 | 0.000000 | 0.007233 | 0.015268 | 0.028169 | 0.095238 |
summary_by_hospital_under_80.to_csv('output/pathway_summary_by_hopsital_under_80.csv')
# Set all StrokeTeam to same
single_group = data_under_80.copy()
single_group['StrokeTeam'] = "na"
analysis_all = analyse_by_team(single_group).T
stats_by_age['under_80'] = analysis_all[0]
Aged 80+#
analysis_by_team_80_plus = analyse_by_team(data_80_plus)
summary_by_hospital_80_plus = analysis_by_team_80_plus.describe().T
summary_by_hospital_80_plus
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| thrombolysis_rate | 132.0 | 0.097072 | 0.037177 | 0.015184 | 0.071957 | 0.090386 | 0.118358 | 0.227273 |
| arrivals | 132.0 | 715.295455 | 324.288269 | 130.000000 | 519.000000 | 693.500000 | 887.250000 | 2119.000000 |
| 80_plus | 132.0 | 1.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| rankin_before_stroke_all | 132.0 | 1.582028 | 0.329839 | 0.799699 | 1.355713 | 1.583800 | 1.794138 | 2.464387 |
| nihss_arrival_all | 132.0 | 8.949314 | 1.154855 | 5.969027 | 8.166105 | 8.990764 | 9.807674 | 12.285714 |
| onset_known | 132.0 | 0.646519 | 0.141493 | 0.310705 | 0.543596 | 0.622715 | 0.740186 | 0.992000 |
| known_arrival_within_4hrs | 132.0 | 0.624133 | 0.093277 | 0.312377 | 0.577418 | 0.627499 | 0.683352 | 0.821990 |
| known_arrival_within_4hrs_all | 132.0 | 0.395484 | 0.069188 | 0.194872 | 0.355810 | 0.392473 | 0.442737 | 0.601429 |
| rankin_before_stroke_4hrs | 132.0 | 1.638257 | 0.335931 | 0.863830 | 1.395506 | 1.650321 | 1.864107 | 2.386986 |
| nihss_arrival_4hrs | 132.0 | 10.546849 | 1.371413 | 6.688889 | 9.689962 | 10.640593 | 11.557606 | 13.642157 |
| onset_arrival_mins_mu | 132.0 | 113.772402 | 7.520809 | 88.483516 | 109.819101 | 113.890154 | 119.189322 | 138.846154 |
| onset_arrival_mins_sigma | 132.0 | 52.245901 | 3.034150 | 45.279315 | 50.524577 | 52.190029 | 53.860620 | 67.187195 |
| scan_within_4_hrs | 132.0 | 0.945029 | 0.032080 | 0.828221 | 0.929246 | 0.947741 | 0.966349 | 1.000000 |
| arrival_scan_mins_mu | 132.0 | 43.433208 | 12.746320 | 15.293680 | 33.818753 | 43.487091 | 51.689702 | 90.817518 |
| arrival_scan_mins_sigma | 132.0 | 42.925626 | 8.063562 | 22.024223 | 38.179966 | 42.849548 | 48.226897 | 62.732866 |
| eligable | 132.0 | 0.253801 | 0.083254 | 0.047297 | 0.200000 | 0.245084 | 0.312157 | 0.523077 |
| scan_needle_mins_mu | 132.0 | 38.902157 | 13.191973 | 15.808989 | 29.224436 | 36.542609 | 44.672101 | 89.818182 |
| scan_needle_mins_sigma | 132.0 | 24.271719 | 9.058096 | 7.718740 | 19.062118 | 22.987181 | 27.544448 | 63.516781 |
| arrival_needle_mins_mu | 132.0 | 61.413428 | 16.096518 | 26.550000 | 51.892105 | 58.293308 | 68.292676 | 117.476190 |
| arrival_needle_mins_sigma | 132.0 | 29.143830 | 9.245836 | 8.274724 | 23.842572 | 27.826304 | 32.099727 | 69.019784 |
| onset_needle_mins_mu | 132.0 | 157.229189 | 15.530805 | 120.300000 | 148.107065 | 154.422078 | 165.543783 | 198.809524 |
| onset_needle_mins_sigma | 132.0 | 49.224555 | 8.075909 | 25.569296 | 44.276242 | 49.327607 | 52.665629 | 82.577511 |
| thrombolysis_after_180_min | 132.0 | 0.300602 | 0.126670 | 0.000000 | 0.222222 | 0.274205 | 0.362623 | 0.681818 |
| thrombolysis_after_270_min | 132.0 | 0.015137 | 0.020425 | 0.000000 | 0.000000 | 0.007547 | 0.026059 | 0.095238 |
summary_by_hospital_80_plus.to_csv('output/pathway_summary_by_hopsital_80_plus.csv')
# Set all StrokeTeam to same
single_group = data_80_plus.copy()
single_group['StrokeTeam'] = "na"
analysis_all = analyse_by_team(single_group).T
stats_by_age['80+'] = analysis_all[0]
Show comparison of means by age group
stats_by_age.to_csv('output/summary_by_age_group.csv')
stats_by_age
| all | under_80 | 80+ | |
|---|---|---|---|
| stroke_team | na | na | na |
| thrombolysis_rate | 0.118239 | 0.130445 | 0.101124 |
| arrivals | 226820 | 132401 | 94419 |
| 80_plus | 0.423219 | 0.0 | 1.0 |
| rankin_before_stroke_all | 1.047315 | 0.654225 | 1.598534 |
| nihss_arrival_all | 7.46584 | 6.363859 | 9.059266 |
| onset_known | 0.668596 | 0.681943 | 0.64988 |
| known_arrival_within_4hrs | 0.586399 | 0.568081 | 0.613354 |
| known_arrival_within_4hrs_all | 0.392064 | 0.387399 | 0.398606 |
| rankin_before_stroke_4hrs | 1.059689 | 0.623704 | 1.653869 |
| nihss_arrival_4hrs | 9.214728 | 8.178815 | 10.66594 |
| onset_arrival_mins_mu | 111.392722 | 109.258422 | 114.30144 |
| onset_arrival_mins_sigma | 52.729071 | 52.840638 | 52.437656 |
| scan_within_4_hrs | 0.94789 | 0.949953 | 0.945079 |
| arrival_scan_mins_mu | 42.101407 | 41.584525 | 42.809469 |
| arrival_scan_mins_sigma | 44.793795 | 44.568632 | 45.091427 |
| eligable | 0.311363 | 0.346537 | 0.263179 |
| scan_needle_mins_mu | 35.955574 | 36.509091 | 34.957163 |
| scan_needle_mins_sigma | 28.380323 | 29.290013 | 26.633541 |
| arrival_needle_mins_mu | 58.816315 | 60.017234 | 56.650144 |
| arrival_needle_mins_sigma | 35.162372 | 36.656941 | 32.180851 |
| onset_needle_mins_mu | 155.77768 | 156.465147 | 154.537656 |
| onset_needle_mins_sigma | 54.035606 | 55.238587 | 51.774937 |
| thrombolysis_after_180_min | 0.295016 | 0.303109 | 0.280419 |
| thrombolysis_after_270_min | 0.016726 | 0.0183 | 0.013887 |
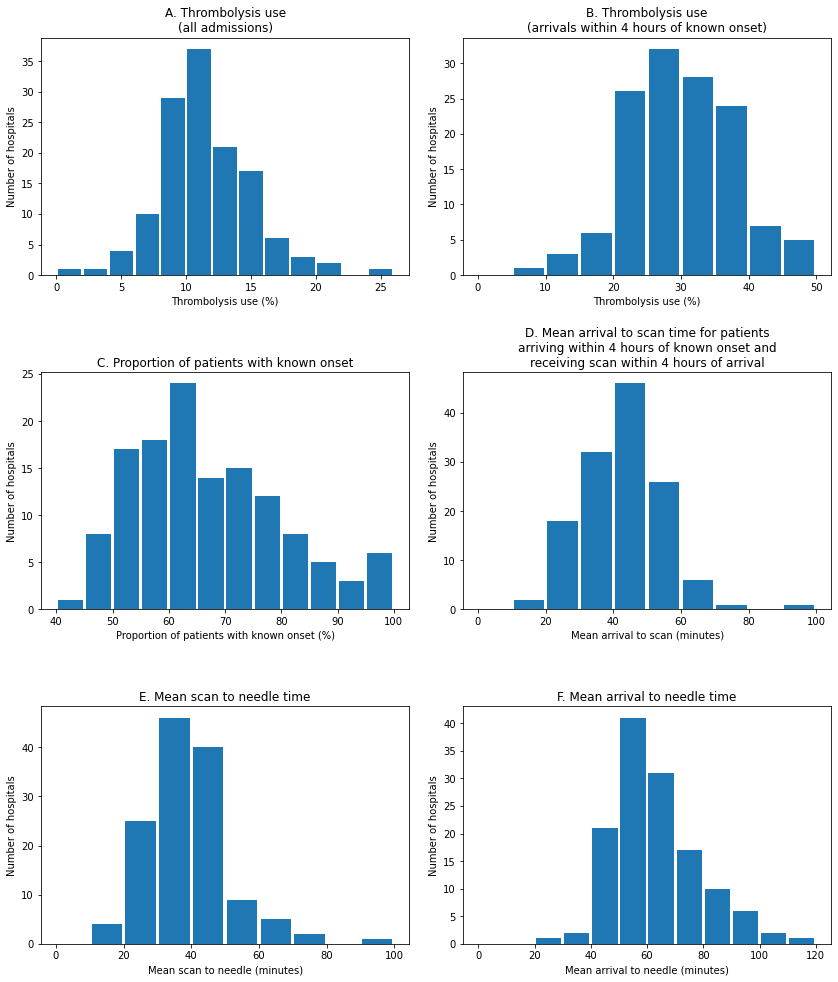
Summary figure#
# Set up figure
fig = plt.figure(figsize=(12,14))
# Subplot 1: Histogram of thrombolysis rate
ax1 = fig.add_subplot(321)
x = analysis_by_team['thrombolysis_rate'] * 100
bins = np.arange(0, x.max()+2, 2)
ax1.hist(x, bins=bins, rwidth=0.9)
ax1.set_xlabel('Thrombolysis use (%)')
ax1.set_ylabel('Number of hospitals')
ax1.set_title('A. Thrombolysis use\n(all admissions)')
# Subplot 2: Histogram of thrombolysis rate for patients arriving in 4 hours
ax2 = fig.add_subplot(322)
x = analysis_by_team['eligable'] * 100
bins = np.arange(0, x.max()+5, 5)
ax2.hist(x, bins=bins, rwidth=0.9)
ax2.set_xlabel('Thrombolysis use (%)')
ax2.set_ylabel('Number of hospitals')
ax2.set_title('B. Thrombolysis use\n(arrivals within 4 hours of known onset)')
# Subplot 3: Histogram of proportion of patients with known onset
ax3 = fig.add_subplot(323)
x = analysis_by_team['onset_known'] * 100
bins = np.arange(40, x.max()+5, 5)
ax3.hist(x, bins=bins, rwidth=0.9)
ax3.set_xlabel('Proportion of patients with known onset (%)')
ax3.set_ylabel('Number of hospitals')
ax3.set_title('C. Proportion of patients with known onset')
# Subplot 4: Histogram of mean Arrival to scan times
ax4 = fig.add_subplot(324)
x = analysis_by_team['arrival_scan_mins_mu']
bins = np.arange(0, x.max()+10, 10)
ax4.hist(x, bins=bins, rwidth=0.9)
ax4.set_xlabel('Mean arrival to scan (minutes)')
ax4.set_ylabel('Number of hospitals')
ax4.set_title('''D. Mean arrival to scan time for patients
arriving within 4 hours of known onset and
receiving scan within 4 hours of arrival''')
# Subplot 5: Histogram of mean scan to needle times
ax5 = fig.add_subplot(325)
x = analysis_by_team['scan_needle_mins_mu']
bins = np.arange(0, x.max()+10, 10)
ax5.hist(x, bins=bins, rwidth=0.9)
ax5.set_xlabel('Mean scan to needle (minutes)')
ax5.set_ylabel('Number of hospitals')
ax5.set_title('E. Mean scan to needle time')
# Subplot 6: Histogram of mean arrival to needle times
ax6 = fig.add_subplot(326)
x = analysis_by_team['arrival_needle_mins_mu']
bins = np.arange(0, x.max()+10, 10)
ax6.hist(x, bins=bins, rwidth=0.9)
ax6.set_xlabel('Mean arrival to needle (minutes)')
ax6.set_ylabel('Number of hospitals')
ax6.set_title('F. Mean arrival to needle time')
# Save and show
plt.tight_layout(pad=2)
plt.savefig('output/pathway_summary_six_histograms.jpg', dpi=300)
plt.show();
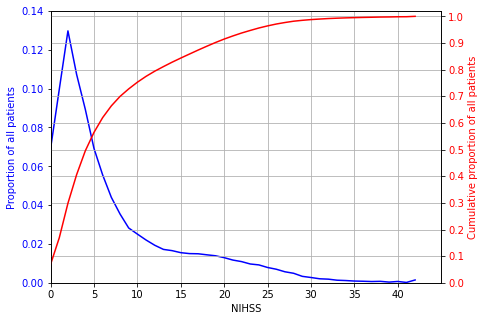
Analysis of stroke severity#
# Restrict to known NIHSS
mask = data['S2NihssArrival'] >= 0
known_NIHSS = data[mask]
counts = pd.DataFrame(known_NIHSS['S2NihssArrival'].value_counts().sort_index())
counts['cum_sum'] = np.cumsum(counts['S2NihssArrival'])
counts
| S2NihssArrival | cum_sum | |
|---|---|---|
| 0.0 | 14658 | 14658 |
| 1.0 | 21117 | 35775 |
| 2.0 | 27521 | 63296 |
| 3.0 | 22765 | 86061 |
| 4.0 | 18955 | 105016 |
| 5.0 | 14686 | 119702 |
| 6.0 | 11783 | 131485 |
| 7.0 | 9334 | 140819 |
| 8.0 | 7519 | 148338 |
| 9.0 | 5987 | 154325 |
| 10.0 | 5322 | 159647 |
| 11.0 | 4675 | 164322 |
| 12.0 | 4102 | 168424 |
| 13.0 | 3649 | 172073 |
| 14.0 | 3513 | 175586 |
| 15.0 | 3292 | 178878 |
| 16.0 | 3190 | 182068 |
| 17.0 | 3171 | 185239 |
| 18.0 | 3058 | 188297 |
| 19.0 | 2953 | 191250 |
| 20.0 | 2747 | 193997 |
| 21.0 | 2484 | 196481 |
| 22.0 | 2316 | 198797 |
| 23.0 | 2056 | 200853 |
| 24.0 | 1955 | 202808 |
| 25.0 | 1673 | 204481 |
| 26.0 | 1483 | 205964 |
| 27.0 | 1204 | 207168 |
| 28.0 | 1040 | 208208 |
| 29.0 | 705 | 208913 |
| 30.0 | 579 | 209492 |
| 31.0 | 439 | 209931 |
| 32.0 | 398 | 210329 |
| 33.0 | 284 | 210613 |
| 34.0 | 247 | 210860 |
| 35.0 | 191 | 211051 |
| 36.0 | 169 | 211220 |
| 37.0 | 139 | 211359 |
| 38.0 | 159 | 211518 |
| 39.0 | 77 | 211595 |
| 40.0 | 146 | 211741 |
| 41.0 | 40 | 211781 |
| 42.0 | 311 | 212092 |
# Set up figure
fig = plt.figure(figsize=(7,5))
ax1 = fig.add_subplot(111)
total = counts['S2NihssArrival'].sum()
x = list(counts.index)
y1 = counts['S2NihssArrival'] / total
y2 = counts['cum_sum'] / total
ax1.plot(x,y1,color='b')
ax1.tick_params(axis='y', labelcolor='b')
ax1.set_ylim(0, 0.14)
ax1.set_ylabel('Proportion of all patients', color='b')
ax1.set_xlabel('NIHSS')
ax1.set_xlim(0,45)
ax1.set_xticks(np.arange(0,42,5))
ax1.grid(axis='x')
# Add cumsum to second axis
ax2 = ax1.twinx()
ax2.plot(x,y2,label='Cumulative sum', color='r')
ax2.set_ylim(0,1.02)
ax2.set_yticks(np.arange(0,1.01,0.1))
ax2.set_xticks(np.arange(0,42,5))
ax2.tick_params(axis='y', labelcolor='r')
ax2.set_ylabel('Cumulative proportion of all patients', color='r')
ax2.grid('both')
plt.savefig('output/nihss_count.jpg', dpi=300)
plt.show()
Analyse onset-to-arrival times#
All arrivals
# Count patients with determined NIHSS
total_with_determined_nihss = np.sum(data['S2NihssArrival'] >= 0)
proportion_with_determined_nihss = np.mean(data['S2NihssArrival'] >= 0)
# Restrict to known onset
f = lambda x: x in ['Precise', 'Best estimate']
mask = data['S1OnsetTimeType'].apply(f)
data_known_onset = data[mask]
prop_known_onset_all = mask.mean()
# Get counts of arrival times, up to 12 hours, in 30 min bins
bins = np.arange(0, 721, 30)
arrivals_all, _ = np.histogram(data_known_onset['S1OnsetToArrival_min'], bins)
prop_arrivals_in_12_hours = np.sum(arrivals_all) / mask.sum()
df_all_arrivals = pd.DataFrame()
df_all_arrivals['Arrival bin (upper)'] = bins[1:]
df_all_arrivals.set_index('Arrival bin (upper)', inplace=True)
df_all_arrivals['Counts'] = arrivals_all
df_all_arrivals['Prop known onset'] = arrivals_all / mask.sum()
df_all_arrivals['Cumulatative prop'] = np.cumsum(df_all_arrivals['Prop known onset'])
print('All arrivals:')
print('-------------')
print(f'Proporotion of arrivals with determmined NIHSS: {proportion_with_determined_nihss:0.3}')
print(f'Proportion of arrivals with known stroke onset: {prop_known_onset_all:0.3f}')
print(f'Proportion known stroke onset arriving within 12 hours: {prop_arrivals_in_12_hours:0.3f}')
print (df_all_arrivals)
All arrivals:
-------------
Proporotion of arrivals with determmined NIHSS: 0.935
Proportion of arrivals with known stroke onset: 0.669
Proportion known stroke onset arriving within 12 hours: 0.799
Counts Prop known onset Cumulatative prop
Arrival bin (upper)
30 1819 0.011995 0.011995
60 11283 0.074401 0.086396
90 23536 0.155198 0.241594
120 18806 0.124008 0.365603
150 12402 0.081780 0.447382
180 8791 0.057969 0.505351
210 6731 0.044385 0.549736
240 5283 0.034837 0.584572
270 4344 0.028645 0.613217
300 3653 0.024088 0.637305
330 3133 0.020659 0.657965
360 2767 0.018246 0.676211
390 2486 0.016393 0.692603
420 2109 0.013907 0.706510
450 1968 0.012977 0.719488
480 1690 0.011144 0.730632
510 1599 0.010544 0.741175
540 1454 0.009588 0.750763
570 1376 0.009073 0.759837
600 1235 0.008144 0.767980
630 1202 0.007926 0.775907
660 1163 0.007669 0.783575
690 1136 0.007491 0.791066
720 1158 0.007636 0.798702
Mild stroke (NIHSS <= 5)
# Proportion mild stroke
mild = data['S2NihssArrival'] <=5
prop_mild = mild.sum()/data.shape[0]
print (f'Proportion mild {prop_mild:0.2f}')
Proportion mild 0.53
# Restrict to NIHSS <5
mask = data['S2NihssArrival'] <=5
data_mild = data[mask]
proportion_mild_stroke = mask.sum() / total_with_determined_nihss
# Restrict to known onset
f = lambda x: x in ['Precise', 'Best estimate']
mask = data_mild['S1OnsetTimeType'].apply(f)
data_known_onset = data_mild[mask]
prop_known_onset_mild = mask.mean()
# Get counts of arrival times, up to 12 hours, in 30 min bins
bins = np.arange(0, 721, 30)
arrivals_all, _ = np.histogram(data_known_onset['S1OnsetToArrival_min'], bins)
prop_arrivals_in_12_hours = np.sum(arrivals_all) / mask.sum()
df_mild_arrivals = pd.DataFrame()
df_mild_arrivals['Arrival bin (upper)'] = bins[1:]
df_mild_arrivals.set_index('Arrival bin (upper)', inplace=True)
df_mild_arrivals['Counts'] = arrivals_all
df_mild_arrivals['Prop known onset'] = arrivals_all / mask.sum()
df_mild_arrivals['Cumulatative prop'] = np.cumsum(df_mild_arrivals['Prop known onset'])
print('Mild stroke (NIHSS <= 5):')
print('-------------------------')
print(f'Proportion of all arrivals, with determined NIHSS, with mild stroke: {proportion_mild_stroke:0.3f}')
print(f'Proportion of arrivals with known stroke onset: {prop_known_onset_mild:0.3f}')
print(f'Proportion known stroke onset arriving within 12 hours: {prop_arrivals_in_12_hours:0.3f}')
print (df_mild_arrivals)
Mild stroke (NIHSS <= 5):
-------------------------
Proportion of all arrivals, with determined NIHSS, with mild stroke: 0.564
Proportion of arrivals with known stroke onset: 0.645
Proportion known stroke onset arriving within 12 hours: 0.723
Counts Prop known onset Cumulatative prop
Arrival bin (upper)
30 810 0.010485 0.010485
60 3767 0.048760 0.059245
90 8347 0.108043 0.167288
120 7769 0.100562 0.267850
150 5840 0.075593 0.343443
180 4290 0.055530 0.398972
210 3464 0.044838 0.443810
240 2807 0.036334 0.480144
270 2369 0.030664 0.510808
300 2010 0.026017 0.536826
330 1760 0.022781 0.559607
360 1545 0.019998 0.579605
390 1409 0.018238 0.597844
420 1240 0.016051 0.613894
450 1204 0.015585 0.629479
480 975 0.012620 0.642099
510 978 0.012659 0.654758
540 895 0.011585 0.666343
570 843 0.010912 0.677255
600 759 0.009824 0.687079
630 743 0.009617 0.696697
660 697 0.009022 0.705719
690 681 0.008815 0.714533
720 682 0.008828 0.723361
Non-mild stroke (NIHSS > 5)
# Proportion non-mild stroke
non_mild = data['S2NihssArrival'] >5
prop_non_mild = non_mild.sum()/data.shape[0]
print (f'Proportion non-mild {prop_non_mild:0.2f}')
Proportion non-mild 0.41
# Restrict to NIHSS >5
mask = data['S2NihssArrival'] >5
data_not_mild = data[mask]
proportion_not_mild_stroke = mask.sum() / total_with_determined_nihss
# Restrict to known onset
f = lambda x: x in ['Precise', 'Best estimate']
mask = data_not_mild['S1OnsetTimeType'].apply(f)
data_known_onset = data_not_mild[mask]
prop_known_onset_not_mild = mask.mean()
# Get counts of arrival times, up to 12 hours, in 30 min bins
bins = np.arange(0, 721, 30)
arrivals_all, _ = np.histogram(data_known_onset['S1OnsetToArrival_min'], bins)
prop_arrivals_in_12_hours = np.sum(arrivals_all) / mask.sum()
df_not_mild_arrivals = pd.DataFrame()
df_not_mild_arrivals['Arrival bin (upper)'] = bins[1:]
df_not_mild_arrivals.set_index('Arrival bin (upper)', inplace=True)
df_not_mild_arrivals['Counts'] = arrivals_all
df_not_mild_arrivals['Prop known onset'] = arrivals_all / mask.sum()
df_not_mild_arrivals['Cumulatative prop'] = np.cumsum(
df_not_mild_arrivals['Prop known onset'])
print('Non-mild stroke (NIHSS >5):')
print('-------------------------')
print(f'Proportion of arrivals with known stroke onset: {prop_known_onset_not_mild:0.3f}')
print(f'Proportion of all arrivals, with determined NIHSS, with NIHSS >5: {proportion_not_mild_stroke:0.3f}')
print(f'Proportion known stroke onset arriving within 12 hours: {prop_arrivals_in_12_hours:0.3f}')
print (df_not_mild_arrivals)
Non-mild stroke (NIHSS >5):
-------------------------
Proportion of arrivals with known stroke onset: 0.715
Proportion of all arrivals, with determined NIHSS, with NIHSS >5: 0.436
Proportion known stroke onset arriving within 12 hours: 0.883
Counts Prop known onset Cumulatative prop
Arrival bin (upper)
30 915 0.013852 0.013852
60 6813 0.103141 0.116993
90 13655 0.206722 0.323715
120 9899 0.149860 0.473575
150 5874 0.088926 0.562501
180 4058 0.061434 0.623935
210 2920 0.044206 0.668140
240 2238 0.033881 0.702021
270 1756 0.026584 0.728605
300 1421 0.021512 0.750117
330 1220 0.018469 0.768587
360 1067 0.016153 0.784740
390 949 0.014367 0.799107
420 750 0.011354 0.810461
450 654 0.009901 0.820362
480 631 0.009553 0.829914
510 537 0.008130 0.838044
540 473 0.007161 0.845205
570 465 0.007040 0.852244
600 406 0.006146 0.858391
630 394 0.005965 0.864355
660 398 0.006025 0.870381
690 396 0.005995 0.876376
720 413 0.006252 0.882628
NIHSS >10
# Restrict to NIHSS >10
mask = data['S2NihssArrival'] >10
data_nihss_11plus = data[mask]
proportion__11plus = mask.sum() / total_with_determined_nihss
# Restrict to known onset
f = lambda x: x in ['Precise', 'Best estimate']
mask = data_nihss_11plus['S1OnsetTimeType'].apply(f)
data_known_onset = data_nihss_11plus[mask]
prop_known_onset_11plus = mask.mean()
# Get counts of arrival times, up to 12 hours, in 30 min bins
bins = np.arange(0, 721, 30)
arrivals_all, _ = np.histogram(data_known_onset['S1OnsetToArrival_min'], bins)
prop_arrivals_in_12_hours = np.sum(arrivals_all) / mask.sum()
df_arrivals_11plus = pd.DataFrame()
df_arrivals_11plus['Arrival bin (upper)'] = bins[1:]
df_arrivals_11plus.set_index('Arrival bin (upper)', inplace=True)
df_arrivals_11plus['Counts'] = arrivals_all
df_arrivals_11plus['Prop known onset'] = arrivals_all / mask.sum()
df_arrivals_11plus['Cumulatative prop'] = np.cumsum(
df_arrivals_11plus['Prop known onset'])
print('Non-mild stroke (NIHSS >10):')
print('-------------------------')
print(f'Proportion of arrivals with known stroke onset: {prop_known_onset_11plus:0.3f}')
print(f'Proportion of all arrivals, with determined NIHSS, with NIHH >10: {proportion__11plus:0.3f}')
print(f'Proportion known stroke onset arriving within 12 hours: {prop_arrivals_in_12_hours:0.3f}')
print (df_arrivals_11plus)
Non-mild stroke (NIHSS >10):
-------------------------
Proportion of arrivals with known stroke onset: 0.726
Proportion of all arrivals, with determined NIHSS, with NIHH >10: 0.247
Proportion known stroke onset arriving within 12 hours: 0.910
Counts Prop known onset Cumulatative prop
Arrival bin (upper)
30 543 0.014264 0.014264
60 4644 0.121992 0.136256
90 8890 0.233529 0.369786
120 6076 0.159609 0.529395
150 3409 0.089550 0.618945
180 2281 0.059919 0.678864
210 1586 0.041662 0.720526
240 1222 0.032100 0.752627
270 951 0.024982 0.777608
300 739 0.019413 0.797021
330 641 0.016838 0.813859
360 549 0.014422 0.828281
390 466 0.012241 0.840522
420 357 0.009378 0.849900
450 305 0.008012 0.857912
480 326 0.008564 0.866476
510 276 0.007250 0.873726
540 228 0.005989 0.879715
570 208 0.005464 0.885179
600 190 0.004991 0.890170
630 173 0.004544 0.894715
660 192 0.005044 0.899758
690 188 0.004939 0.904697
720 197 0.005175 0.909872
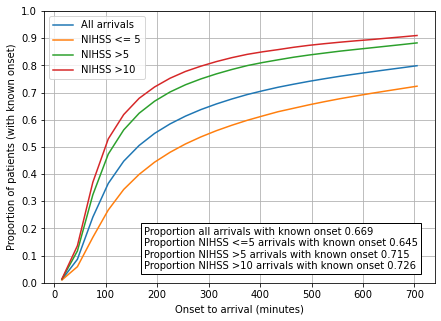
# Set up figure
fig = plt.figure(figsize=(7,5))
ax = fig.add_subplot(111)
x = df_all_arrivals.index
x -= 15 # plot at centre of bin
y1 = df_all_arrivals['Cumulatative prop']
y2 = df_mild_arrivals['Cumulatative prop']
y3 = df_not_mild_arrivals['Cumulatative prop']
y4 = df_arrivals_11plus['Cumulatative prop']
ax.plot(x, y1, label = 'All arrivals')
ax.plot(x, y2, label = 'NIHSS <= 5')
ax.plot(x, y3, label = 'NIHSS >5')
ax.plot(x, y4, label = 'NIHSS >10')
ax.set_xlabel('Onset to arrival (minutes)')
ax.set_ylabel('Proportion of patients (with known onset)')
ax.set_ylim(0, 720)
ax.set_ylim(0, 1.0)
ax.set_yticks(np.arange(0, 1.01, 0.1))
ax.grid()
text = f'''Proportion all arrivals with known onset {prop_known_onset_all:0.3}
Proportion NIHSS <=5 arrivals with known onset {prop_known_onset_mild:0.3}
Proportion NIHSS >5 arrivals with known onset {prop_known_onset_not_mild:0.3}
Proportion NIHSS >10 arrivals with known onset {prop_known_onset_11plus:0.3}'''
props = dict(boxstyle='square', facecolor='white', alpha=1.0)
plt.text(175, 0.05, text, bbox=props)
ax.legend(loc='upper left')
plt.savefig('output/onset_to_arrival.jpg', dpi=300)
plt.show()
Queries on time of arrival and NIHSS for patients who receive thrombolysis#
See also later section on comparison of average values for patients who receive thrombolysis and those that do not (though that data is for patients who arrive within 4 hours of known stroke onset)
data_thrombolysed = data[data['S2Thrombolysis'] == 'Yes']
thrombolysed = dict()
# Onset known
onset_known = (data_thrombolysed['S1OnsetTimeType'] == 'Precise') | (data_thrombolysed['S1OnsetTimeType'] == 'Best estimate')
thrombolysed['onset known'] = onset_known.mean()
# Arrival within 4 hours
arrive_4_hours = data_thrombolysed['S1OnsetToArrival_min'] <= 240
thrombolysed['arrive within 4 hours'] = arrive_4_hours.mean()
# Arrival within 6 hours
arrive_6_hours = data_thrombolysed['S1OnsetToArrival_min'] <= 360
thrombolysed['arrive within 6 hours'] = arrive_6_hours.mean()
# NIHSS >5
nihss_greater_5 = data_thrombolysed['S2NihssArrival'] >5
thrombolysed['nihss 6+'] = nihss_greater_5.mean()
nihss_greater_5_arrival_4_hrs = nihss_greater_5 & arrive_4_hours
thrombolysed['nihss 6+ and arrive in 4 hours'] = nihss_greater_5_arrival_4_hrs.mean()
nihss_greater_5_arrival_6_hrs = nihss_greater_5 & arrive_6_hours
thrombolysed['nihss 6+ and arrive in 6 hours'] = nihss_greater_5_arrival_6_hrs.mean()
# NIHSS >10
nihss_greater_10 = data_thrombolysed['S2NihssArrival'] >10
thrombolysed['nihss 11+'] = nihss_greater_10.mean()
nihss_greater_10_arrival_4_hrs = nihss_greater_10 & arrive_4_hours
thrombolysed['nihss 11+ and arrive in 4 hours'] = nihss_greater_10_arrival_4_hrs.mean()
nihss_greater_10_arrival_6_hrs = nihss_greater_10 & arrive_6_hours
thrombolysed['nihss 11+ and arrive in 6 hours'] = nihss_greater_10_arrival_6_hrs.mean()
thrombolysed = pd.Series(thrombolysed)
thrombolysed
onset known 0.988926
arrive within 4 hours 0.979045
arrive within 6 hours 0.987211
nihss 6+ 0.739252
nihss 6+ and arrive in 4 hours 0.723330
nihss 6+ and arrive in 6 hours 0.729520
nihss 11+ 0.451247
nihss 11+ and arrive in 4 hours 0.440807
nihss 11+ and arrive in 6 hours 0.444573
dtype: float64
General observations#
There are 239,505 admissions (2016-2018) to 132 stroke teams that receive at least 300 admissions and provide thrombolysis to at least 10 patients over the course of three years (2016-2018).
5.3% of patients have an in-hospital stroke onset; 12.3% receive thrombolysis. 94.7% of patients have an out-of-hospital stroke onset; 11.8% receive thrombolysis.
The following statistic apply to out-of-hospital onset stroke patients only
43% of arrivals are aged 80 or more.
67% of all patients have a determined stroke time of onset, 60% of whom arrive within 4 hours of known stroke onset. 40% of all arrivals arrive within 4 hours of known stroke onset.
The average NIHSS (stroke severity) is 7.4
Of those arriving within 4 hours of known stroke onset:
The average onset-to-arrival time is 111 minutes.
95% receive a scan within 4 hours of arrival, with an average arrival-to-scan time of 43 minutes.
30% of those who receive a scan within 4 hours of known stroke onset, and 11.8% of all arrivals, receive thrombolysis. The average scan-to-needle time is 40 minutes, the average arrival-to-needle time is 63 minutes, and the average onset-to-needle time is 158 minutes.
On inter-hospital variation:
Thrombolysis use varies from 1.5% to 24.3% of all patients, and 7.3% to 49.7% of patients arriving within 4 hours of known stroke onset.
The proportion of determined stroke onsets ranges from 34% to 99%.
The proportion of patients arriving within 4 hours of known stroke onset ranges from 22% to 56%.
The proportion of patients scanned within 4 hours of arrival ranges from 85% to 100%.
Average arrival-to-scan (for those arriving within 4 hours of known stroke onset and scanned within 4 hours of arrival) ranges from 19 to 93 minutes.
Average arrival-to-needle time varies from 26 to 111 minutes.
The proportion of patients aged 80+ varies from 29% to 58%.
The average NIHSS (stroke severity) ranges from 6.1 to 11.7.
On differences by age group (under age 80 years vs 80+):
10.1% of arrivals aged 80+ receive thrombolysis, c.f. 13.0% of those under age 80.
39% of arrivals aged 80+ arrive within 4 hours of known stroke onset, c.f. 40% of those under age 80.
The average disability (Rankin) before stroke is 1.7 for those aged 80+, c.f. 0.6 for those under age 80.
The average stroke severity (NIHSS) on arrival is 10.7 for those aged 80+, c.f. 8.2 for those under age 80.
Of those scanned within 4 hours, 26.3% of those aged 80+ receive thrombolysis, c.f. 34.7% of those under age 80.
On onset-to-arrival times
Those with non-mild stroke (NINHSS >5) tend to arrive earlier than mild stroke (NIHSS <=5). For example, of those with known stroke onset time, 70% and 79% of those with non-mild stroke arrive within 4 and 6 hours from onset. The same results for mild stroke patients are 48% and 58%. Non-mild stroke are also more likely to have a determined time of onset compared to mild stroke (72% vs 65%).