Modular TensorFlow model with 2D embedding - Train and save model for 10k patient subset
Contents
Modular TensorFlow model with 2D embedding - Train and save model for 10k patient subset#
Embedding converts a categorical variable into a projection onto n-dimensional space [1], and has been shown to be an effective way to train neural network when using categorical data, while also allowing a measure of similarity/distance between different values of the categorical data, Here we use embedding for hospital ID. We also convert patient data and pathway data into an embedded vector (this may also be known as encoding the data in a vector with fewer dimensions than the original data set for those groups of features).
[1] Guo C, & Berkhahn F. (2016) Entity Embeddings of Categorical Variables. arXiv:160406737 [cs] http://arxiv.org/abs/1604.06737
Aims#
To train and save fully connected models for 10k patient subset
Basic methodology#
Models are fitted to previously split training and test data sets.
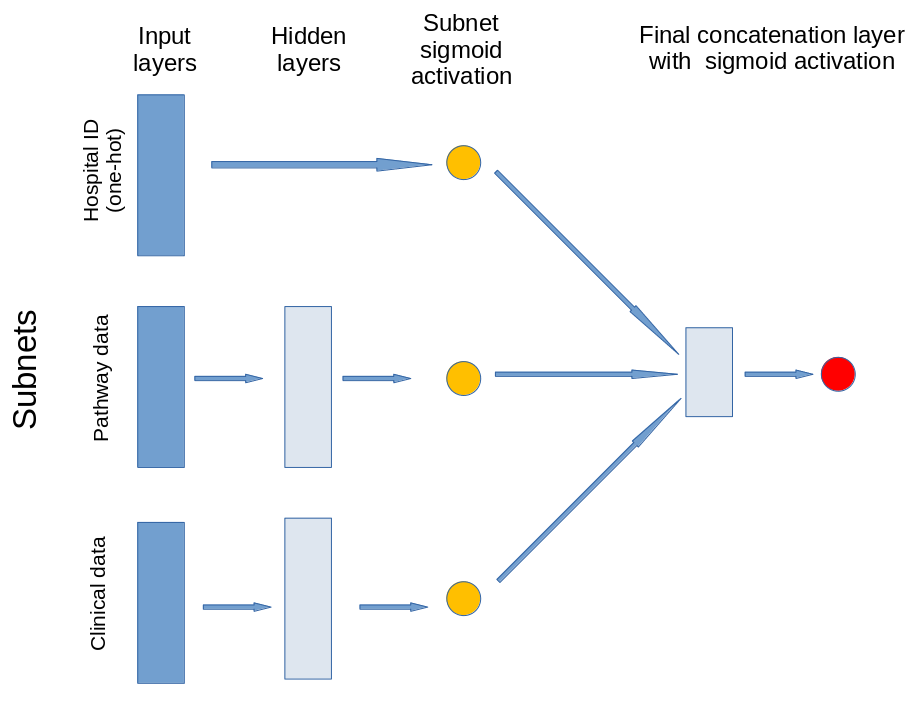
The model contains three subnets that take portions of the data. The output of these subnets is an n-dimensional vector. In this case the output is a 2D vector, that is each subnet is reduced to a single value output. The subnets created are for:
Patient clinical data: Age, gender, ethnicity, disability before stroke, stroke scale data. Pass through one hidden layer (with 2x neurons as input features) and then to single neuron with sigmoid activation.
Pathway process data: Times of arrival and scan, time of day, day of week. Pass through one hidden layer (with 2x neurons as input features) and then to single neuron with sigmoid activation.
Hospital ID (one-hot encoded): Connect input directly to single neuron with sigmoid activation.
The outputs of the three subnet outputs are then passed to a single neuron with sigmoid activation for final output.
Models are fitted with keras ‘early-stopping’. This monitors accuracy of the test set and stops when there has been no improvement in n epochs (this is the ‘patience’, and is set to 100 epochs here).
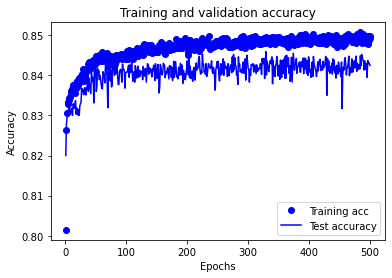
NOTE: In the training charts below you will see that test accuracy is higher than training accuracy. This may seem surprising; it is due to keras returning the accuracy of the training set during training when dropout is used (and no dropout is used for the test set).
Set up#
path = './saved_models/2d_for_10k/'
# Turn warnings off to keep notebook tidy
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
# sklearn for pre-processing
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import StratifiedKFold
# TensorFlow api model
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Model
from tensorflow.keras.optimizers import Adam
from tensorflow.keras import backend as K
from tensorflow.keras.losses import binary_crossentropy
Define function to scale data#
In neural networks it is common to to scale input data 0-1 rather than use standardisation (subtracting mean and dividing by standard deviation) of each feature).
def scale_data(X_train, X_test):
"""Scale data 0-1 based on min and max in training set"""
# Initialise a new scaling object for normalising input data
sc = MinMaxScaler()
# Set up the scaler just on the training set
sc.fit(X_train)
# Apply the scaler to the training and test sets
train_sc = sc.transform(X_train)
test_sc = sc.transform(X_test)
return train_sc, test_sc
Set up neural net#
def make_net(number_features_patient,
number_features_pathway,
number_features_hospital,
patient_encoding_neurones=2,
pathway_encoding_neurones=2,
hospital_encoding_neurones=2,
expansion=2,
learning_rate=0.003,
dropout=0.5):
# Clear Tensorflow
K.clear_session()
# Patient (clinical data) encoding layers
input_patient = layers.Input(shape=number_features_patient)
# Patient dense layer 1
patient_dense_1 = layers.Dense(
number_features_patient * expansion, activation='relu')(input_patient)
patient_norm_1 = layers.BatchNormalization()(patient_dense_1)
patient_dropout_1 = layers.Dropout(dropout)(patient_norm_1)
# Patient encoding layer
patient_encoding = layers.Dense(
patient_encoding_neurones, activation='sigmoid',
name='patient_encode')(patient_dropout_1)
# Pathway encoding layers
input_pathway = layers.Input(shape=number_features_pathway)
# pathway dense layer 1
pathway_dense_1 = layers.Dense(
number_features_pathway * expansion, activation='relu')(input_pathway)
pathway_norm_1 = layers.BatchNormalization()(pathway_dense_1)
pathway_dropout_1 = layers.Dropout(dropout)(pathway_norm_1)
# pathway encoding layer
pathway_encoding = layers.Dense(
pathway_encoding_neurones, activation='sigmoid',
name='pathway_encode')(pathway_dropout_1)
# hospital encoding layers
input_hospital = layers.Input(shape=number_features_hospital)
# hospital encoding layer
hospital_encoding = layers.Dense(
hospital_encoding_neurones, activation='sigmoid',
name='hospital_encode')(input_hospital)
# Concatenation layer
concat = layers.Concatenate()(
[patient_encoding, pathway_encoding, hospital_encoding])
# Outpout (single sigmoid)
outputs = layers.Dense(1, activation='sigmoid')(concat)
# Build net
net = Model(inputs=[
input_patient, input_pathway, input_hospital], outputs=outputs)
# Compiling model
opt = Adam(lr=learning_rate)
net.compile(loss='binary_crossentropy',
optimizer=opt,
metrics=['accuracy'])
return net
Train model#
# Get data subgroups
subgroups = pd.read_csv('../data/subnet.csv', index_col='Item')
# Get list of clinical items
clinical_subgroup = subgroups.loc[subgroups['Subnet']=='clinical']
clinical_subgroup = list(clinical_subgroup.index)
# Get list of pathway items
pathway_subgroup = subgroups.loc[subgroups['Subnet']=='pathway']
pathway_subgroup = list(pathway_subgroup.index)
# Get list of hospital items
hospital_subgroup = subgroups.loc[subgroups['Subnet']=='hospital']
hospital_subgroup = list(hospital_subgroup.index)
# Load data
train = pd.read_csv(f'../data/10k_training_test/cohort_10000_train.csv')
test = pd.read_csv(f'../data/10k_training_test/cohort_10000_test.csv')
# OneHot encode stroke team
coded = pd.get_dummies(train['StrokeTeam'])
train = pd.concat([train, coded], axis=1)
train.drop('StrokeTeam', inplace=True, axis=1)
coded = pd.get_dummies(test['StrokeTeam'])
test = pd.concat([test, coded], axis=1)
test.drop('StrokeTeam', inplace=True, axis=1)
# Split into X, y
X_train_df = train.drop('S2Thrombolysis',axis=1)
y_train_df = train['S2Thrombolysis']
X_test_df = test.drop('S2Thrombolysis',axis=1)
y_test_df = test['S2Thrombolysis']
# Split train and test data by subgroups
X_train_patients = X_train_df[clinical_subgroup]
X_test_patients = X_test_df[clinical_subgroup]
X_train_pathway = X_train_df[pathway_subgroup]
X_test_pathway = X_test_df[pathway_subgroup]
X_train_hospitals = X_train_df[hospital_subgroup]
X_test_hospitals = X_test_df[hospital_subgroup]
# Convert to NumPy
X_train = X_train_df.values
X_test = X_test_df.values
y_train = y_train_df.values
y_test = y_test_df.values
# Scale data
X_train_patients_sc, X_test_patients_sc = \
scale_data(X_train_patients, X_test_patients)
X_train_pathway_sc, X_test_pathway_sc = \
scale_data(X_train_pathway, X_test_pathway)
X_train_hospitals_sc, X_test_hospitals_sc = \
scale_data(X_train_hospitals, X_test_hospitals)
# Define network
number_features_patient = X_train_patients_sc.shape[1]
number_features_pathway = X_train_pathway_sc.shape[1]
number_features_hospital = X_train_hospitals_sc.shape[1]
model = make_net(
number_features_patient,
number_features_pathway,
number_features_hospital)
# Define save checkpoint callback (only save if new best validation results)
checkpoint_cb = keras.callbacks.ModelCheckpoint(
'model_checkpoint_2d.h5', save_best_only=True)
# Define early stopping callback: Stop when no validation improvement
# Restore weights to best validation accuracy
early_stopping_cb = keras.callbacks.EarlyStopping(
patience=100, restore_best_weights=True)
# Train model (including class weights)
history = model.fit(
[X_train_patients_sc, X_train_pathway_sc, X_train_hospitals_sc],
y_train,
epochs=500,
batch_size=32,
validation_data=(
[X_test_patients_sc, X_test_pathway_sc, X_test_hospitals_sc],
y_test),
verbose=0)
#callbacks=[checkpoint_cb, early_stopping_cb])
### Test model
probability = model.predict(
[X_train_patients_sc, X_train_pathway_sc, X_train_hospitals_sc])
y_pred_train = probability >= 0.5
y_pred_train = y_pred_train.flatten()
accuracy_train = np.mean(y_pred_train == y_train)
print(f'Accuracy train {accuracy_train:0.3f}', end=' ')
probability = model.predict(
[X_test_patients_sc, X_test_pathway_sc, X_test_hospitals_sc])
y_pred_test = probability >= 0.5
y_pred_test = y_pred_test.flatten()
accuracy_test = np.mean(y_pred_test == y_test)
print(f'Accuracy test {accuracy_test:0.3f}')
# save model
filename = f'{path}model.h5'
model.save(filename);
# Plot training accuracy
history_dict = history.history
acc_values = history_dict['accuracy']
val_acc_values = history_dict['val_accuracy']
epochs = range(1, len(acc_values) + 1)
plt.plot(epochs, acc_values, 'bo', label='Training acc')
plt.plot(epochs, val_acc_values, 'b', label='Test accuracy')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
Accuracy train 0.858 Accuracy test 0.843