Fully connected TensorFlow model - Check required training epochs
Contents
Fully connected TensorFlow model - Check required training epochs#
Aims#
Ascertain how many training epochs are needed to train a fully connected TensorFlow neural net. This will be used in future training.
Basic methodology#
Models are fitted to previously split training and test data sets.
MinMax scaling is used (all features are scaled 0-1 based on the feature min/max).
Model has two hidden layers, each with the number of neurons being 2x the number of features. Prior studies show performance of the network is similar across all models with this complexity or more. A dropout value of 0.5 is used based on previous exploration.
A batch size of 32 is used (“Friends don’t let friends use mini-batches larger than 32”. Yan LeCun on paper: arxiv.org/abs/1804.07612)
Model structure:
Input layer
Dense layer (# neurons = 2x features, ReLu activation)
Batch normalisation
Dropout layer
Dense layer (# neurons = 2x features, ReLu activation)
Batch normalisation
Dropout layer
Output layer (single sigmoid activation)
# Turn warnings off to keep notebook tidy
import warnings
warnings.filterwarnings("ignore")
Load modules#
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
# sklearn for pre-processing
from sklearn.preprocessing import MinMaxScaler
# TensorFlow api model
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Model
from tensorflow.keras.optimizers import Adam
from tensorflow.keras import backend as K
from tensorflow.keras.losses import binary_crossentropy
Function to scale data (minmax scaling)#
def scale_data(X_train, X_test):
"""Scale data 0-1 based on min and max in training set"""
# Initialise a new scaling object for normalising input data
sc = MinMaxScaler()
# Set up the scaler just on the training set
sc.fit(X_train)
# Apply the scaler to the training and test sets
train_sc = sc.transform(X_train)
test_sc = sc.transform(X_test)
return train_sc, test_sc
Define neural net#
def make_net(number_features, expansion=2, learning_rate=0.003, dropout=0.5):
# Clear Tensorflow
K.clear_session()
# Input layer
inputs = layers.Input(shape=number_features)
# Dense layer 1
dense_1 = layers.Dense(
number_features * expansion, activation='relu')(inputs)
norm_1 = layers.BatchNormalization()(dense_1)
dropout_1 = layers.Dropout(dropout)(norm_1)
# Dense layer 2
dense_2 = layers.Dense(
number_features * expansion, activation='relu')(dropout_1)
norm_2 = layers.BatchNormalization()(dense_2)
dropout_2 = layers.Dropout(dropout)(norm_2)
# Outpout (single sigmoid)
outputs = layers.Dense(1, activation='sigmoid')(dropout_2)
# Build net
net = Model(inputs, outputs)
# Compiling model
opt = Adam(lr=learning_rate)
net.compile(loss='binary_crossentropy',
optimizer=opt,
metrics=['accuracy'])
return net
Fit models#
# Loop through 5 k-folds
test_acc = []
for k in range(5):
# Load data
train = pd.read_csv(f'../data/kfold_5fold/train_{k}.csv')
test = pd.read_csv(f'../data/kfold_5fold/test_{k}.csv')
# OneHot encode stroke team
coded = pd.get_dummies(train['StrokeTeam'])
train = pd.concat([train, coded], axis=1)
train.drop('StrokeTeam', inplace=True, axis=1)
coded = pd.get_dummies(test['StrokeTeam'])
test = pd.concat([test, coded], axis=1)
test.drop('StrokeTeam', inplace=True, axis=1)
# Split into X, y
X_train_df = train.drop('S2Thrombolysis',axis=1)
y_train_df = train['S2Thrombolysis']
X_test_df = test.drop('S2Thrombolysis',axis=1)
y_test_df = test['S2Thrombolysis']
# Convert to NumPy
X_train = X_train_df.values
X_test = X_test_df.values
y_train = y_train_df.values
y_test = y_test_df.values
# Scale data
X_train_sc, X_test_sc = scale_data(X_train, X_test)
# Define network
number_features = X_train_sc.shape[1]
model = make_net(number_features)
# Define save checkpoint callback (only save if new best validation results)
checkpoint_cb = keras.callbacks.ModelCheckpoint(
'model_checkpoint.h5', save_best_only=True)
# Define early stopping callback: Stop when no validation improvement
# Restore weights to best validation accuracy
early_stopping_cb = keras.callbacks.EarlyStopping(
patience=50, restore_best_weights=True)
# Train model (including class weights)
history = model.fit(X_train_sc,
y_train,
epochs=100,
batch_size=32,
validation_data=(X_test_sc, y_test),
verbose=0)
# callbacks=[checkpoint_cb, early_stopping_cb]
### Test model
probability = model.predict(X_train_sc)
y_pred_train = probability >= 0.5
y_pred_train = y_pred_train.flatten()
accuracy_train = np.mean(y_pred_train == y_train)
print(f'Model {k}: Accuracy train {accuracy_train:0.3f}', end=' ')
probability = model.predict(X_test_sc)
y_pred_test = probability >= 0.5
y_pred_test = y_pred_test.flatten()
accuracy_test = np.mean(y_pred_test == y_test)
print(f'Accuracy test {accuracy_test:0.3f}')
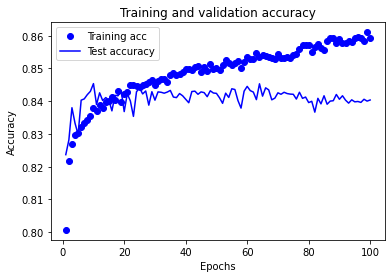
# Plot training accuracy
history_dict = history.history
acc_values = history_dict['accuracy']
val_acc_values = history_dict['val_accuracy']
test_acc.append(val_acc_values)
epochs = range(1, len(acc_values) + 1)
plt.plot(epochs, acc_values, 'bo', label='Training acc')
plt.plot(epochs, val_acc_values, 'b', label='Test accuracy')
plt.title('Training and validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
# Remove model
del model
Model 0: Accuracy train 0.878 Accuracy test 0.846
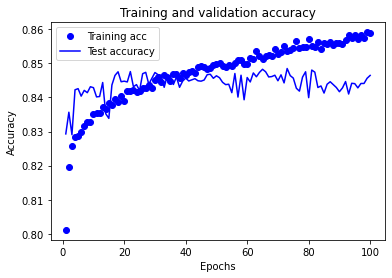
Model 1: Accuracy train 0.879 Accuracy test 0.842
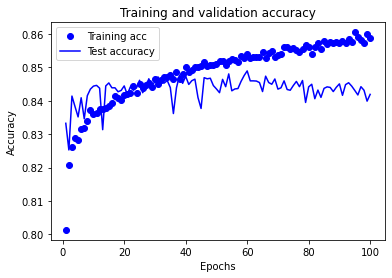
Model 2: Accuracy train 0.873 Accuracy test 0.838
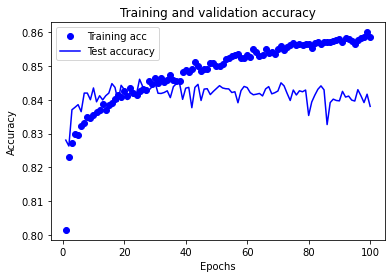
Model 3: Accuracy train 0.875 Accuracy test 0.846
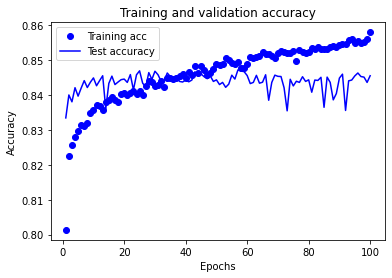
Model 4: Accuracy train 0.874 Accuracy test 0.840
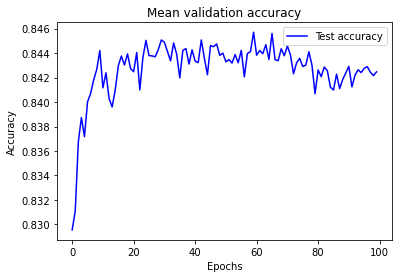
results = np.array(test_acc)
mean_results = results.mean(axis=0)
epochs = range(100)
plt.plot(epochs, mean_results, 'b', label='Test accuracy')
plt.title('Mean validation accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
Observations#
Training set accuracy continues to increase throughout the training period of the experiment.
Test set accuracy rises, plateaus, and then declines over the training period of the experiment. The plateau occurs between approximately 25 and 70 epochs.
A training period of 30 epochs (mini-batch size 32) may be used in future experiments using this model structure.