Pathway patterns throughout the week in an example single stroke team
Contents
Pathway patterns throughout the week in an example single stroke team#
Aims#
Show key pathway statistics broken down by day of week in a single example team.
Import libraries and data#
Data has been restricted to stroke teams with at least 300 admissions, with at least 10 patients receiving thrombolysis, over three years.
# import libraries
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
# Display entire dataframes
pd.set_option("display.max_rows", 999, "display.max_columns", 150)
# import data
raw_data = pd.read_csv(
'./../data/2019-11-04-HQIP303-Exeter_MA.csv', low_memory=False)
headings = list(raw_data)
print (headings)
['StrokeTeam', 'PatientUID', 'Pathway', 'S1AgeOnArrival', 'MoreEqual80y', 'S1Gender', 'S1Ethnicity', 'S1OnsetInHospital', 'S1OnsetToArrival_min', 'S1OnsetDateType', 'S1OnsetTimeType', 'S1ArriveByAmbulance', 'S1AdmissionHour', 'S1AdmissionDay', 'S1AdmissionQuarter', 'S1AdmissionYear', 'CongestiveHeartFailure', 'Hypertension', 'AtrialFibrillation', 'Diabetes', 'StrokeTIA', 'AFAntiplatelet', 'AFAnticoagulent', 'AFAnticoagulentVitK', 'AFAnticoagulentDOAC', 'AFAnticoagulentHeparin', 'S2INR', 'S2INRHigh', 'S2INRNK', 'S2NewAFDiagnosis', 'S2RankinBeforeStroke', 'Loc', 'LocQuestions', 'LocCommands', 'BestGaze', 'Visual', 'FacialPalsy', 'MotorArmLeft', 'MotorArmRight', 'MotorLegLeft', 'MotorLegRight', 'LimbAtaxia', 'Sensory', 'BestLanguage', 'Dysarthria', 'ExtinctionInattention', 'S2NihssArrival', 'S2BrainImagingTime_min', 'S2StrokeType', 'S2Thrombolysis', 'Haemorrhagic', 'TimeWindow', 'Comorbidity', 'Medication', 'Refusal', 'Age', 'Improving', 'TooMildSevere', 'TimeUnknownWakeUp', 'OtherMedical', 'S2ThrombolysisTime_min', 'S2TIAInLastMonth']
Restrict original data to hospitals with at least 300 admissions + 10 thrombolysis patients#
keep = []
groups = raw_data.groupby('StrokeTeam') # creates a new object of groups of data
for index, group_df in groups: # each group has an index and a dataframe of data
# Skip if total admissions less than 300 or total thrombolysis < 10
admissions = group_df.shape[0]
thrombolysis_received = group_df['S2Thrombolysis'] == 'Yes'
if (admissions < 300) or (thrombolysis_received.sum() < 10):
continue
else:
keep.append(group_df)
# Concatenate output
data = pd.DataFrame()
data = pd.concat(keep)
Remove in-hospital onset
mask = data['S1OnsetInHospital'] == 'No'
data = data[mask]
Restrict to single stroke_team#
team_pick = 0
teams = list(set(data['StrokeTeam']))
teams.sort()
team = teams[team_pick]
mask = data['StrokeTeam'] == team
selected_data = data[mask]
print (f'Arrivals {mask.sum()}')
Arrivals 2015
Group by day of week#
# Work on copy of data
data_time = selected_data.copy()
# Set up results DataFrame
day_summary = pd.DataFrame()
# Count arrivals
day_summary['Arrivals'] = \
data_time.groupby('S1AdmissionDay').count()['StrokeTeam']
# Get thrombolysis rate
thrombolysed = data_time['S2Thrombolysis'] == 'Yes'
data_time['thrombolysed'] = thrombolysed
day_summary['thrombolyse_all'] = \
data_time.groupby('S1AdmissionDay').mean()['thrombolysed']
# Get proportion of strokes with known onset
onset_known = (data_time['S1OnsetTimeType'] == 'Best estimate') | \
(data_time['S1OnsetTimeType'] == 'Precise')
data_time['onset_known'] = onset_known
day_summary['onset_known'] = \
data_time.groupby('S1AdmissionDay').mean()['onset_known']
# Get proportion over 80
data_time['age_80_plus'] = data_time['MoreEqual80y'] == 'Yes'
day_summary['age_80_plus'] = \
data_time.groupby('S1AdmissionDay').mean()['age_80_plus']
# Get Rankin score
day_summary['rankin_all'] = \
data_time.groupby('S1AdmissionDay').mean()['S2RankinBeforeStroke']
# Get NIHSS
day_summary['nihss_all'] = \
data_time.groupby('S1AdmissionDay').mean()['S2NihssArrival']
# Get onset to arrival <4hrs and then restrict data
data_time['4hr_arrival'] = data_time['S1OnsetToArrival_min'] <= 240
day_summary['4hr_arrival'] = \
data_time.groupby('S1AdmissionDay').mean()['4hr_arrival']
mask = data_time['4hr_arrival']
data_time = data_time[mask]
# Get Rankin score of arrivals within 4hrs onset
day_summary['rankin_4hr'] = \
data_time.groupby('S1AdmissionDay').mean()['S2RankinBeforeStroke']
# Get NIHSS of arrivals within 4hrs onset
day_summary['nihss_4hr'] = \
data_time.groupby('S1AdmissionDay').mean()['S2NihssArrival']
# Get onset to arrival (of those arriving within 4 hours)
day_summary['onset_arrival'] = \
data_time.groupby('S1AdmissionDay').mean()['S1OnsetToArrival_min']
# Get scan in four hours (and remove rest)
data_time['4hr_scan'] = data_time['S2BrainImagingTime_min'] <= 240
day_summary['scan_4hrs'] = \
data_time.groupby('S1AdmissionDay').mean()['4hr_scan']
mask = data_time['4hr_scan']
data_time = data_time[mask]
# Get arrival to scan (of those arriving within 4 hours and scanned in 4hrs)
day_summary['arrival_scan'] = \
data_time.groupby('S1AdmissionDay').mean()['S2BrainImagingTime_min']
# Filter down to acanned within 4 hrs onset
onset_to_scan = (
data_time['S1OnsetToArrival_min'] + data_time['S2BrainImagingTime_min'])
data_time['onset_to_scan'] = onset_to_scan
mask = data_time['onset_to_scan'] <= 240
data_time = data_time[mask]
# Get thrombolysis given and remove rest
day_summary['thrombolyse_4hr'] = \
data_time.groupby('S1AdmissionDay').mean()['thrombolysed']
mask = data_time['thrombolysed']
# Get scan to needle
scan_to_needle = (
data_time['S2ThrombolysisTime_min'] - data_time['S2BrainImagingTime_min'])
data_time['scan_needle'] = scan_to_needle
day_summary['scan_to_needle'] = \
data_time.groupby('S1AdmissionDay').mean()['scan_needle']
# Get arrival to needle
day_summary['arrival_to_needle'] = \
data_time.groupby('S1AdmissionDay').mean()['S2ThrombolysisTime_min']
# Get onset to needle
onset_to_needle = (
data_time['S1OnsetToArrival_min'] + data_time['S2ThrombolysisTime_min'])
data_time['onset_to_needle'] = onset_to_needle
day_summary['onset_to_needle'] = \
data_time.groupby('S1AdmissionDay').mean()['onset_to_needle']
# Sort by day
day_order = ['Monday', 'Tuesday', 'Wednesday', 'Thursday', 'Friday', 'Saturday',
'Sunday']
day_summary = day_summary.loc[day_order]
day_summary['Weekday'] = ['Mo', 'Tu', 'We', 'Th', 'Fr', 'Sa', 'Su']
Show summary table by day of week#
day_summary
| Arrivals | thrombolyse_all | onset_known | age_80_plus | rankin_all | nihss_all | 4hr_arrival | rankin_4hr | nihss_4hr | onset_arrival | scan_4hrs | arrival_scan | thrombolyse_4hr | scan_to_needle | arrival_to_needle | onset_to_needle | Weekday | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1AdmissionDay | |||||||||||||||||
| Monday | 306 | 0.163399 | 0.620915 | 0.447712 | 1.052288 | 8.879310 | 0.388889 | 0.899160 | 10.412281 | 109.369748 | 0.966387 | 19.000000 | 0.449541 | 43.285714 | 53.326531 | 148.040816 | Mo |
| Tuesday | 277 | 0.173285 | 0.657040 | 0.425993 | 0.870036 | 9.337121 | 0.451264 | 0.800000 | 12.049587 | 106.520000 | 0.984000 | 20.609756 | 0.417391 | 42.916667 | 55.062500 | 147.104167 | Tu |
| Wednesday | 272 | 0.169118 | 0.621324 | 0.463235 | 1.198529 | 7.855469 | 0.455882 | 1.016129 | 9.264463 | 114.225806 | 0.943548 | 12.410256 | 0.394737 | 41.155556 | 47.488889 | 149.555556 | We |
| Thursday | 306 | 0.147059 | 0.633987 | 0.428105 | 1.111111 | 8.802768 | 0.408497 | 0.928000 | 10.785124 | 112.016000 | 0.976000 | 25.139344 | 0.403670 | 46.840909 | 55.272727 | 145.772727 | Th |
| Friday | 335 | 0.152239 | 0.608955 | 0.438806 | 1.035821 | 9.586319 | 0.438806 | 1.081633 | 10.791367 | 113.088435 | 0.979592 | 17.354167 | 0.360294 | 39.142857 | 46.530612 | 139.163265 | Fr |
| Saturday | 259 | 0.142857 | 0.648649 | 0.432432 | 0.918919 | 8.794979 | 0.432432 | 0.892857 | 9.271028 | 110.437500 | 0.982143 | 21.609091 | 0.377551 | 56.324324 | 64.783784 | 160.324324 | Sa |
| Sunday | 260 | 0.134615 | 0.665385 | 0.450000 | 0.934615 | 8.445833 | 0.461538 | 0.950000 | 10.324074 | 109.933333 | 0.925000 | 14.198198 | 0.317757 | 62.058824 | 68.441176 | 176.205882 | Su |
Show summary charts of key metrics by day of week#
# Set up figure
fig = plt.figure(figsize=(12,15))
# Subplot 1: Arrivals
ax1 = fig.add_subplot(4,4,1)
x = day_summary['Weekday']
y = day_summary['Arrivals'] / day_summary['Arrivals'].sum() * 7
ax1.plot(x,y)
# Add line at 1
y1 = np.repeat(1,7)
ax1.plot(x,y1, color='0.5', linestyle=':')
ax1.set_ylim(ymin=0) # must be after plot method
ax1.set_xlabel('Day of week')
ax1.set_ylabel('Normlalised arrivals')
ax1.set_title('Arrivals\n(normalised to average)')
# Subplot 2: Thrombolysis
ax2 = fig.add_subplot(4,4,2)
x = day_summary['Weekday']
y = day_summary['thrombolyse_all'] * 100
ax2.plot(x,y)
ax2.set_xlabel('Day of week')
ax2.set_ylabel('Thrombolysis (%)')
ax2.set_title('Thrombolysis use\n(all arrivals)')
# Subplot 3: Known onset
ax3 = fig.add_subplot(4,4,3)
x = day_summary['Weekday']
y = day_summary['onset_known'] * 100
ax3.plot(x,y)
ax3.set_xlabel('Day of week')
ax3.set_ylabel('Onset known (%)')
ax3.set_title('Proportion with known onset')
# Subplot 4: age_80_plus
ax4 = fig.add_subplot(4,4,4)
x = day_summary['Weekday']
y = day_summary['age_80_plus'] * 100
ax4.plot(x,y)
ax4.set_xlabel('Day of week')
ax4.set_ylabel('Aged 80+')
ax4.set_title('Proportion aged 80+')
# Subplot 5: Rankin (all arrivals)
ax5 = fig.add_subplot(4,4,5)
x = day_summary['Weekday']
y = day_summary['rankin_all']
ax5.plot(x,y)
ax5.set_xlabel('Day of week')
ax5.set_ylabel('Rankin (mean)')
ax5.set_title('Mean pre-stroke\nmodified Rankin\n(all arrivals)')
# Subplot 6: NIHSS (all arrivals)
ax6 = fig.add_subplot(4,4,6)
x = day_summary['Weekday']
y = day_summary['nihss_all']
ax6.plot(x,y)
ax6.set_xlabel('Day of week')
ax6.set_ylabel('NIHSS (mean)')
ax6.set_title('Mean NIH Stroke Scale\n(all arrivals)')
# Subplot 7: 4hr_arrival
ax7 = fig.add_subplot(4,4,7)
x = day_summary['Weekday']
y = day_summary['4hr_arrival'] * 100
ax7.plot(x,y)
ax7.set_xlabel('Day of week')
ax7.set_ylabel('Arrive within 4hrs of onset (%)')
ax7.set_title('Proportion arriving within\n4hrs of known onset')
# Subplot 8: Rankin (4hr arrivals)
ax8 = fig.add_subplot(4,4,8)
x = day_summary['Weekday']
y = day_summary['rankin_4hr']
ax8.plot(x,y)
ax8.set_xlabel('Day of week')
ax8.set_ylabel('Rankin (mean)')
ax8.set_title('Mean pre-stroke\nmodified Rankin\n(arrivals 4hrs from onset)')
# Subplot 9: NIHSS (4hr arrivals)
ax9 = fig.add_subplot(4,4,9)
x = day_summary['Weekday']
y = day_summary['nihss_4hr']
ax9.plot(x,y)
ax9.set_xlabel('Day of week')
ax9.set_ylabel('NIHSS (mean)')
ax9.set_title('Mean NIH Stroke Scale\n(arrivals 4hrs from onset)')
# Subplot 10: onset_arrival (4hr arrivals)
ax10 = fig.add_subplot(4,4,10)
x = day_summary['Weekday']
y = day_summary['onset_arrival']
ax10.plot(x,y)
ax10.set_xlabel('Day of week')
ax10.set_ylabel('Onset to arrival (minutes, mean)')
ax10.set_title('Mean onset to arrival\n(arrivals 4hrs from onset)')
# Subplot 11: scan_4hrs (4hr arrivals)
ax11 = fig.add_subplot(4,4,11)
x = day_summary['Weekday']
y = day_summary['scan_4hrs'] * 100
ax11.plot(x,y)
ax11.set_xlabel('Day of week')
ax11.set_ylabel('Proportion scanned within 4hrs (%)')
ax11.set_title('Proportion scanned within\n4hrs of arrival\n(arrivals 4hrs from onset)')
# Subplot 12: arrival_scan (4hr scan)
ax12 = fig.add_subplot(4,4,12)
x = day_summary['Weekday']
y = day_summary['arrival_scan']
ax12.plot(x,y)
ax12.set_xlabel('Day of week')
ax12.set_ylabel('Arrival to scan (minutes, mean)')
ax12.set_title('Mean arrival to scan\n(scanned 4hrs from onset)')
# Subplot 13: thrombolysis (4hr scan)
ax13 = fig.add_subplot(4,4,13)
x = day_summary['Weekday']
y = day_summary['thrombolyse_4hr'] * 100
ax13.plot(x,y)
ax13.set_xlabel('Day of week')
ax13.set_ylabel('Thrombolsyis(%)')
ax13.set_title('Thrombolysis use\n(scanned 4hrs from onset)')
# Subplot 14: scan_to_needle
ax14 = fig.add_subplot(4,4,14)
x = day_summary['Weekday']
y = day_summary['scan_to_needle']
ax14.plot(x,y)
ax14.set_xlabel('Day of week')
ax14.set_ylabel('Scan to needle (minutes, mean)')
ax14.set_title('Mean scan to needle')
# Subplot 15: arrival_to_needle
ax15 = fig.add_subplot(4,4,15)
x = day_summary['Weekday']
y = day_summary['arrival_to_needle']
ax15.plot(x,y)
ax15.set_xlabel('Day of week')
ax15.set_ylabel('Arrival to needle (minutes, mean)')
ax15.set_title('Mean arrival to needle')
# Subplot 16: onset_to_needle
ax16 = fig.add_subplot(4,4,16)
x = day_summary['Weekday']
y = day_summary['onset_to_needle']
ax16.plot(x,y)
ax16.set_xlabel('Day of week')
ax16.set_ylabel('Onset to needle (minutes, mean)')
ax16.set_title('Mean onset to needle')
# Save and show
plt.tight_layout(pad=2)
plt.savefig('output/stats_by_day_of_week_single_team.jpg', dpi=300)
plt.show();
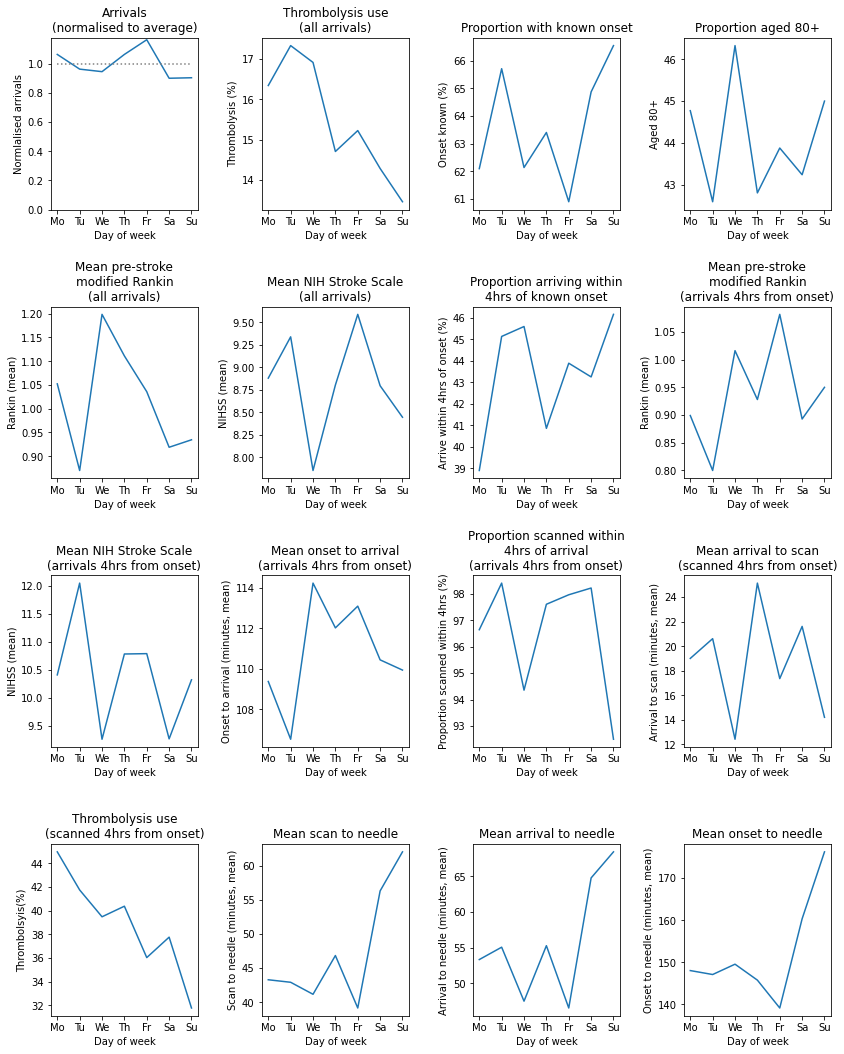
Observations#
Individual hospitals may show similarities and differences in diurnal patterns to national average.
This hospital shows a marked reduction in use of thrombolysis at weekends, which is associated with a lower use of thrombolysis in those scanned within 4 hours and a significantly slower scan-to-needle times. These observations may suggest that this unit struggles maintaining the post-scan thrombolysis pathway at weekends.